Leaf-cutting ants (LCA) are dominant herbivores in the Neotropical rain and dry forests and savannas, removing up to 30% of green biomass in the foraging area of a colony (Siqueira et al. Reference Siqueira, Ribeiro Neto, Tabarelli, Wirth, Andersen and Leal2018, Urbas et al. Reference Urbas, Araújo, Leal and Wirth2007, Wirth et al. Reference Wirth, Herz, Ryel, Beyschlag and Hölldobler2003). In human-modified landscapes their impact is amplified by the fact that LCA colonies proliferate in disturbed habitats such as early-successional forests (Vasconcelos & Cherrett Reference Vasconcelos and Cherrett1995), forest remnants (Terborgh et al. Reference Terborgh, Lopez, Nunez, Rao, Shahabuddin, Orihuela, Riveros, Ascanio, Adler, Lambert and Balbas2001) and forest edges (Wirth et al. Reference Wirth, Meyer, Almeida, Araújo, Barbosa and Leal2007). Understanding LCA diet in disturbed forests could help us to predict their success and impacts on the ecosystem.
Although LCA are largely polyphagous and able to harvest up to 50% of local plant species (Wirth et al. Reference Wirth, Herz, Ryel, Beyschlag and Hölldobler2003), they do exhibit a pronounced food-plant selection. Laboratory experiments have demonstrated that LCA prefer younger and softer leaves over harder and older ones (Howard Reference Howard1988, Nichols-Orians & Schultz Reference Nichols-Orians and Schultz1989). In field studies, LCA were shown to prefer particular plant groups such as the light-demanding pioneer species, presumably due to softer leaves (i.e. reduced morphological defences) and fewer chemical defences associated with a fast-growth strategy (Falcão et al. Reference Falcão, Pinto, Wirth and Leal2011, Farji-Brener Reference Farji-Brener2001, Wirth et al. Reference Wirth, Herz, Ryel, Beyschlag and Hölldobler2003).
In this perspective, research in human-modified tropical landscapes, particularly in fragmented forests, can provide insights into the patterns of LCA diet because pioneer plant species become more abundant in edge-affected habitats (Laurance et al. Reference Laurance, Nascimento, Laurance, Andrade, Fearnside, Ribeiro and Capretz2006, Santos et al. Reference Santos, Peres, Oliveira, Grillo, Alves-Costa and Tabarelli2008), leading to reduced species diversity (Tabarelli et al. Reference Tabarelli, Lopes and Peres2008). Although not all fragmented forests lose tree phylogenetic diversity (Arroyo-Rodríguez et al. Reference Arroyo-Rodríguez, Cavender-Bares, Escobar, Melo, Tabarelli and Santos2012, Matos et al. Reference Matos, Magnago, Gastauer, Carreiras, Simonelli, Meira-Neto and Edwards2017), pioneer species tend to be more related than late-successional species and drive the phylogenetic impoverishment of the forest (Norden et al. Reference Norden, Letcher, Boukili, Swenson and Chazdon2012, Santos et al. Reference Santos, Arroyo-Rodríguez, Moreno and Tabarelli2010, Reference Santos, Tabarelli, Melo, Camargo, Andrade, Laurance and Laurance2014). While phylogenetic distance between neighbouring plant species is likely to interfere with the ability of herbivores to exploit their food plants (Yguel et al. Reference Yguel, Bailey, Tosh, Vialatte, Vasseur, Vitrac, Jean and Prinzing2011), the effect of such human-induced floristic shifts on LCA diet remains entirely unstudied.
Here we examine LCA diet composition in a fragmented Atlantic forest. We hypothesize that: (1) There is a phylogenetic signal in LCA diet following the preference for pioneer species; (2) LCA diet consists of more abundant plant species; (3) LCA diet consists of species with less physical resistance such as softer leaves; and (4) LCA diet depends on the forest habitat (interior vs. edge).
We conducted the study in the Coimbra forest (8°30′S, 35°50′W), the largest (3500 ha) and best-preserved fragment of Atlantic rain forest in north-east Brazil (Santos et al. Reference Santos, Peres, Oliveira, Grillo, Alves-Costa and Tabarelli2008). Coimbra is located on a low-altitude plateau (300–400 m asl). Annual rainfall is ∼2000 mm, with a 3-mo dry season (< 60 mm mo−1) from November to January. The most species-rich families in the region are Fabaceae, Lauraceae, Sapotaceae, Chrysobalanaceae and Lecythidaceae (Santos et al. Reference Santos, Peres, Oliveira, Grillo, Alves-Costa and Tabarelli2008, Reference Santos, Arroyo-Rodríguez, Moreno and Tabarelli2010).
We studied food plant selection, i.e. presence/absence of plant species in the diet (hereafter: diet) of five adult colonies of Atta cephalotes (L.) at the edge (<100 m from forest border, sensu Laurance et al. Reference Laurance, Ferreira, Rankin-de Merona and Laurance1998) and five in the interior (>200 m from forest border) of Coimbra forest. The inter-colony distances were 2.0 ± 1.4 km (mean ± SD) in forest edge and 1.1 ± 0.4 km in the interior. The colonies were studied in every second month over a period of 1 y, and lists of plant species attacked by each colony were compiled (Falcão et al. Reference Falcão, Pinto, Wirth and Leal2011). The data from individual colonies were used as independent data points in each habitat.
We used full woody-plant species lists of the edge and interior habitats of Coimbra forest (Santos et al. Reference Santos, Arroyo-Rodríguez, Moreno and Tabarelli2010) as a reference species pool to infer the diet of A. cephalotes, and to obtain data on the relative abundance of plant species in the two habitats. To test if ant diet consists of softer leaves, we gathered data from the TRY Plant Trait database on leaf mechanical resistance, reflected by leaf tensile strength in N/m (Kattge et al. Reference Kattge, Diaz, Lavorel, Prentice, Leadley, Bönisch, Garnier, Westoby, Reich, Wright and Cornelissen2011). When species-level data were not available, we used the genus-level means. When genus-level data were not available (N = 48 of 175 species) we used the mean value of our species list (the results remained highly significant when species with missing data were excluded).
We constructed a time-calibrated phylogeny for the full plant species list of Coimbra forest. For every species we searched for four DNA regions (matK, 5.8S, rbcL, trnL-trnF) from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). For most species not all sequences were available, or individual species were identified to the genus level only (N = 50), thus we used sequences of congeneric species. We aligned the sequences separately and concatenated them in a supermatrix in Geneious 9.1.3 (www.geneious.com). We used the program BEAST v1.8.0 (Drummond et al. Reference Drummond, Suchard, Xie and Rambaut2012) for the Bayesian analysis and to include time calibration for the tree. To time calibrate the tree we adopted four calibration points (Myrtales, Malpighiales, Sapindales and Eudicotyledoneae; Magallón et al. Reference Magallón, Gómez-Acevedo, Sánchez-Reyes and Hernández-Hernández2015).
We analysed phylogenetic signal in the diet of A. cephalotes with D-value for discrete traits (Fritz & Purvis Reference Fritz and Purvis2010). We also tested for phylogenetic signal in leaf mechanical resistance and in plant species abundance in forest habitats with Pagel’s λ-value for continuous traits (Pagel Reference Pagel1999). These analyses were done using the R packages ‘caper’ and ‘phytools’. We performed phylogenetic generalized linear models (phyloglm) to test the effect of leaf mechanical resistance and plant species abundance at forest edge and interior on ant diet in these habitats, taking into account plant phylogeny to control for the effect of species’ relatedness on trait values. These analyses were conducted in R using the packages ‘car’ and ‘phylolm’.
There was a phylogenetic signal in the diet of A. cephalotes (D = 0.52, P(random) < 0.0001, P(Brownian) < 0.0001; Figure 1). The ants selected both ancient (e.g. Piperaceae, Lauraceae) and recent (e.g. Malpighiaceae, Euphorbiaceae) plant groups, with most species belonging to Malpighiales, Rubiaceae and Melastomataceae (Figure 1). We also found a phylogenetic signal in leaf mechanical resistance (λ = 0.25, P(random) = 0.0006, P(Brownian) < 0.0001), but species abundance in the habitats did not show a phylogenetic signal. The results of the phylogenetic models show that irrespective of phylogeny, ant food species have less resistant leaves at both forest edges and interior (edge: z = −3.2, P = 0.0342; interior: z = −3.6, P = 0.0004). Moreover, more abundant species in the forest were only part of the diet of A. cephalotes at the forest edge (edge: z = 2.1, P = 00342; interior: z = 1.2, P = ns).
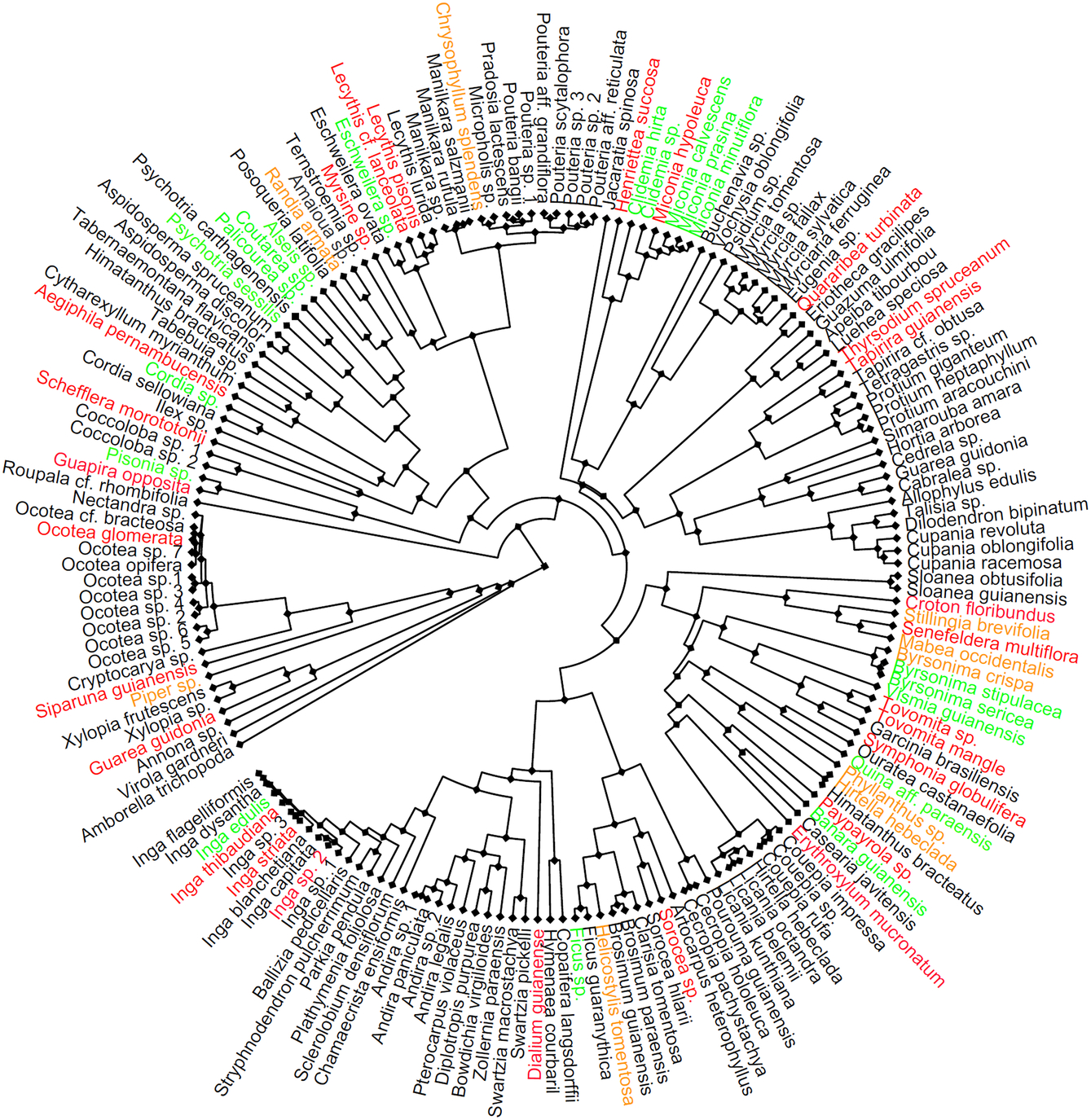
Figure 1. Phylogenetic tree of plant species that occur at the edge and in the interior of Atlantic forest in north-east Brazil (8°30′S, 35°50′W). Coloured names indicate species present in leaf-cutting ant (Atta cephalotes) diet, with phylogenetic signal (D = 0.52; P(random) < 0.0001, P(Brownian) < 0.0001). Orange = species in ant diet in forest interior, green = species in ant diet at forest edge, red = species in ant diet in the two habitats.
Our study confirms that LCA diet is clearly not random. Firstly, our results suggest that the diet is concentrated on some phylogenetic groups, i.e. there is indeed a phylogenetic signal in the diet. Secondly, we found that irrespective of phylogeny, A. cephalotes strongly select food plants with mechanically less resistant leaves both at forest edge and interior. Additionally, our results revealed phylogenetic signal in leaf resistance, which means that less resistant leaves are more frequent across particular plant clades. Thus, leaf mechanical resistance represents an explanation for why the ants are specialized to specific clades. However, while not addressed in this study, this does not exclude the potential influence of other leaf traits, such as plant secondary compounds for LCA diet selection (Wirth et al. Reference Wirth, Herz, Ryel, Beyschlag and Hölldobler2003). Moreover, using abundance data on LCA food plant selection (rather than the presence/absence data available to us) may provide additional insights into the nature of LCA diet preference.
The phylogenetic signal in the diet of A. cephalotes could be caused by their selection of pioneer species as food plants. Although it is not uniformly established whether pioneer species belong to phylogenetically closely related clades in neotropical forests (Norden et al. Reference Norden, Letcher, Boukili, Swenson and Chazdon2012, Santos et al. Reference Santos, Arroyo-Rodríguez, Moreno and Tabarelli2010, Reference Santos, Tabarelli, Melo, Camargo, Andrade, Laurance and Laurance2014), our results suggest this relationship due to the phylogenetic signal in ant diet, and because in the studied forest pioneer species account for more than a half of both tree species and individuals in edge-affected habitats (Oliveira et al. Reference Oliveira, Grillo and Tabarelli2004, Santos et al. Reference Santos, Peres, Oliveira, Grillo, Alves-Costa and Tabarelli2008). We also found that A. cephalotes uses plant species that are more abundant in forest edges but not in forest interiors. The ability of herbivores to exploit their food plants has been found to increase with the phylogenetic proximity between neighbouring plant species (Yguel et al. Reference Yguel, Bailey, Tosh, Vialatte, Vasseur, Vitrac, Jean and Prinzing2011). Our results confirm these findings because of the phylogenetic signal in ant diet and because of the abundance of pioneer species in forest edges. In order to understand the possible reciprocal influence of diet abundance and LCA colony establishment, we encourage future studies to investigate the abundance of plant species inside the foraging territories of LCA.
Phylogenetic signal in LCA diet has multifarious implications for biodiversity persistence in human-modified forests. The existence or lack of phylogenetic signal in pioneer species results in various effects on vegetation. When pioneer species are more related than late-successional species, their proliferation drives phylogenetic impoverishment of the forest (Norden et al. Reference Norden, Letcher, Boukili, Swenson and Chazdon2012, Santos et al. Reference Santos, Arroyo-Rodríguez, Moreno and Tabarelli2010, Reference Santos, Tabarelli, Melo, Camargo, Andrade, Laurance and Laurance2014). However, this is not always the case because not all fragmented forests lose tree phylogenetic diversity (Santo-Silva et al. Reference Santo-Silva, Santos, Arroyo-Rodríguez, Melo, Faria, Cazetta, Mariano-Neto, Hernández-Ruedas and Tabarelli2018). We conclude that by favouring or disfavouring specific phylogenetic clades via herbivory, LCA are key ecological players in the conversion of Neotropical old-growth forests into human-modified secondary forests.
Financial support
The initial study was supported by the German Science Foundation (DFG, grant WI 1959/1-1), Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, grant 007/01), and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq, grant 540322/01-6). The study was also supported by the TRY initiative on plant traits (http://www.try-db.org). PG and EMSR were supported by the Estonian Research Council (grant PUT1006), and EMSR was supported by CNPq post-doc grant (PDJ, 165867/2015-9). BAS, MT and IRL thank CNPq for research grants (310340/2016-0, 310228/2016-6 and 305611/2014-3, respectively). PG and JS thank the Estonian Government for continuously keeping up our hopes about raising research funding to 1% of GDP.



