Introduction
Zhukov (Reference Truett and Kieleczawa1971) established the new genus Parasaccocoelium Zhukov, 1971 as a member of Haploporidae Nicoll, 1914. The type species of this genus, Parasaccocoelium mugili Zhukov, 1971, was found in the intestine of Planiliza haematocheila (Temminck & Schlegel, 1845) in the Japan Sea Basin (Zhukov, Reference Truett and Kieleczawa1971). Overstreet & Curran (Reference Overstreet, Curran, Gibson, Jones and Bray2005) decided that the genus Parasaccocoelium Zhukov, 1971 was invalid and transferred a single species of this genus, P. mugili, to the genus Pseudohapladena Yamaguti, 1952. Later, on the basis of morphological and molecular data, the validity of the genus Parasaccocoelium and the type species P. mugili was confirmed, and two new species, Parasaccocoelium haematocheilum Besprozvannykh, Atopkin, Ermolenko & Nikitenko, 2015 and Parasaccocoelium polyovum Besprozvannykh, Atopkin, Ermolenko & Nikitenko, 2015 (Besprozvannykh et al., Reference Besprozvannykh, Atopkin, Ermolenko and Nikitenko2015) were described from intestines of mullet from south of the Russian Far East.
Concerning Haplosplanchnoidea Poche, 1926, most of the representatives of this subfamily were detected in mullet from the Indo-Western Pacific (Madhavi, Reference Nahhas, Rhodes and Seeto2005), as was the case for Parasaccocoelium species. Species identification of most worms in both Haplosplanchnus Looss, 1902 and other genera of the Haplosplanchnidae Poche, 1926 were based only on morphometric data. Until now, molecular data have been available for a few species only (Cribb et al., Reference Cribb, Bray, Littlewood, Pichelin, Herniou, Littlewood and Bray2001; Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003; Besprozvannykh et al., Reference Besprozvannykh, Atopkin, Ngo, Ermolenko, Ha, Tang and Nikitenko2016; Huston et al., Reference Huston, Cutmore and Cribb2017, Reference Huston, Cutmore and Cribb2018). In Reference Huston, Cutmore and Cribb2018, Huston et al. established the new genus Trigonocephalotrema Huston, Cutmore & Cribb, 2018 with three new species, which were included into the Haplosplanchnidae on the basis of morphological and molecular characteristics. Differentiation at molecular level between Trigonocephalotrema and other representatives of the Haplosplanchnidae required the erection of a new subfamily for this new genus. Given the low number of species within this family for which morphological and molecular data are available, the authors found this insufficient for haplosplanchnid systematics and retained the generic status of Trigonocephalotrema trematodes, further proposing to avoid the concept of subfamilies for the Haplosplanchnidae.
In the present study, we provide morphological and molecular data for a new species of the genus Parasaccocoelium collected from Mugil cephalus from south of the Far East of Russia, and also for worms of a new genus of Haplosplosplanchninae Poche, 1926 and we present molecular data for Hymenocotta mulli collected from Moolgarda seheli off the coast of Vietnam.
Material and methods
Collection of trematodes
Adult worms were collected from the intestines of mullet fish (Mugilidae) from coastal waters of the Primorsky region of the south of the Russian Far East and Cat Ba Island, Vietnam. Worms from the fish, previously defined under a microscope, were rinsed in saline, killed in hot distilled water and preserved in 70% ethanol. After fixation, they were replaced in 96% ethanol. Whole-mounts were made by staining specimens with alum carmine, dehydrating the worms in graded ethanol series and clearing in clove oil. The clove oil treatment was followed by mounting the specimens in Canada balsam under a coverslip on a glass slide. All measurements are given in micrometres.
DNA extraction, amplification and sequencing
Two adult specimens of Parasaccocoelium armatum n. sp., five specimens of Pseudohaplosplanchnus catbaensis n. g. n. sp. and two specimens of H. mulli from 96% ethanol were used for molecular analysis (table 1). Total DNA was extracted from flukes using a ‘hot shot’ technique (Truett, Reference Zhukov2006).
Table 1. List of taxa incorporated in the molecular analysis of the family Haploporidae, with the number of DNA sequences given in parentheses.

Nuclear 18S ribosomal DNA (rDNA) and 28S rDNA fragments were successfully amplified using polymerase chain reaction (PCR). Then, 18S rDNA was amplified with the primers 18S-E (5′ CCG AAT TCG ACA ACC TGG TTG ATC CTG CCA GT 3′) and 18S-F (5′ CCA GCT TGA TCC TTC TGC AGG TTC ACC TAC 3′), as described earlier (Littlewood & Olson, Reference Littlewood, Olson, Littlewood and Bray2001). Initial PCR reaction was performed in a total volume of 20 μl containing 0.25 mm of each primer pair, 25 ng of total DNA in water, 5× Taq buffer, 1.25 mm dNTPs, 1.5 mm magnesium and one unit of Taq polymerase. Amplification of a 2000-bp fragment of 18S rRNA gene was performed in a GeneAmp 9700 (Applied Biosystems, Waltham, Massachusetts, USA), with a 5-min denaturation at 96°C, 35 cycles of 1 min at 96°C, 20 s at 58°C and 5 min at 72°C, and a 10-min extension at 72°C. Negative and positive controls using both primers were used.
The 28S rDNA was amplified with the primers DIG12 (5′-AAG CAT ATC ACT AAG CGG-3′) and 1500R (5′-GCT ATC CTG AGG GAA ACT TCG-3′) (Tkach et al., Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003) with an annealing temperature of 55°C. PCR products were directly sequenced using an ABI Big Dye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, Massachusetts, USA), as recommended by the manufacturer, with the internal sequencing primers described by Tkach et al. (Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003) for 28S rDNA. PCR product sequences were analysed using an ABI 3130 genetic analyser (Applied Biosystems, Waltham, Massachusetts, USA) at the Federal Scientific Center of the East Asia Terrestrial Biodiversity FEB RAS. Sequences were submitted to the GenBank database (National Center for Biotechnology Information (NCBI)).
Alignments and phylogenetic analysis
rDNA sequences were assembled with SeqScape v.2.6 software, provided by Applied Biosystems (Waltham, Massachusetts, USA). Alignments and estimations of the number of variable sites and sequence differences were performed using the MEGA 7.0 software (Kumar et al., Reference Kumar, Stecher and Tamura2016). The values of genetic p-distances were calculated for the 28S rDNA fragment. Phylogenetic relationships were obtained using a concatenated data set of the complete 18S rRNA gene and partial sequences of the 28S rRNA gene. Phylogenetic analysis was performed using the Bayesian algorithm with the MrBayes version 3.1.2 software (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001). The best nucleotide substitution models – the TIM3 + G (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012) for Waretrematinae and TVM + I+G (Posada, Reference Posada and Baxevanis2003) for Haplosplanchnoidea – were estimated with jModeltest version 2.1.5 software (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). Bayesian analysis was performed using 10,000,000 generations with two independent runs. Summary parameters and the phylogenetic tree were calculated with a burn-in of 25% of generations. The significance of the phylogenetic relationships was estimated using posterior probabilities (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001). GenBank sequence data for representatives of Waretrematinae and Haplosplanchnoidea and outgroup taxa used in molecular analysis, including references and accession numbers, are given in the tables 1 and 2.
Table 2. List of taxa incorporated in the molecular analysis of the superfamily Haplosplanchnoidea, with the number of DNA sequences given in parentheses.

Results
Parasaccocoelium armatum n. sp.
Taxonomic summary
Type host. Mugil cephalus Linnaeus, 1758.
Number of fish examined. 97.
Infection of fish. 1.
Intensity of infection. 17 worms.
Site. Intestine.
Type locality. Primorsky region, Kievka River (42°85′20″N, 13°38′390″E).
Type deposition. Type number 152-Tr, paratype number 153-156-Tr. This material is held in the parasitological collection of the Zoological Museum (Federal Scientific Center of the East Asia Terrestrial Biodiversity Far Eastern Branch of Russian Academy of Sciences, Vladivostok, Russia; e-mail: petrova@ibss.dvo.ru). Deposited: 2019.05.05.
Etymology. Species was named because of presence of arming of hermaphroditic canal.
Description
Based on five specimens (fig. 1a–e; table 3). Body elongated, fusiform with spines from anterior end to ventral sucker. Forebody and posterior end of body capable of retracting into inside of body. Eyespot pigment dispersed in forebody. Oral sucker subterminal. Prepharynx long or short. Pharynx transversally oval. Oesophagus shorter, equal or longer than prepharynx, bifurcating at level of or posterior to posterior margin of ventral sucker. Caeca short, sac-shaped terminate near posterior margin middle third of body. Ventral sucker larger than oral sucker, on border of anterior and middle third of body, or in beginning middle third of body. Testis single, V-shaped or from two equal lobes in posterior third of body. Hermaphroditic sac sac-shaped, extends posteriorly beyond ventral sucker, contains internal seminal vesicle, some prostatic cells and long hermaphroditic canal. Hermaphroditic canal thick-walled, muscular and armed with six rows of pads. Pads with two spines on reticular sclerotized base. External seminal vesicle round, extended to ovary. Genital pore, immediately anterior to ventral sucker. Ovary round or oval anterior to testis. Uterus short, located from hermaphroditic sac up to anterior margin of the testis, containing 2–7 eggs. Uterine seminal receptacle present. Metraterm short, with thin walls. Eggs light yellow, oval, operculate, at various stages of embryogeny. Vitellarium in two lateral fields formed from compact follicles of round forms, extending between posterior half of ventral sucker and testis, and can partly cover ovary and testis. Vitelline fields located diagonally relative to each other. Anterior-edge dextral vitelline field at level of posterior half of ventral sucker. Anterior-edge sinistral vitelline field at level of posterior-end dextral vitelline field. Excretory bladder Y-shaped.
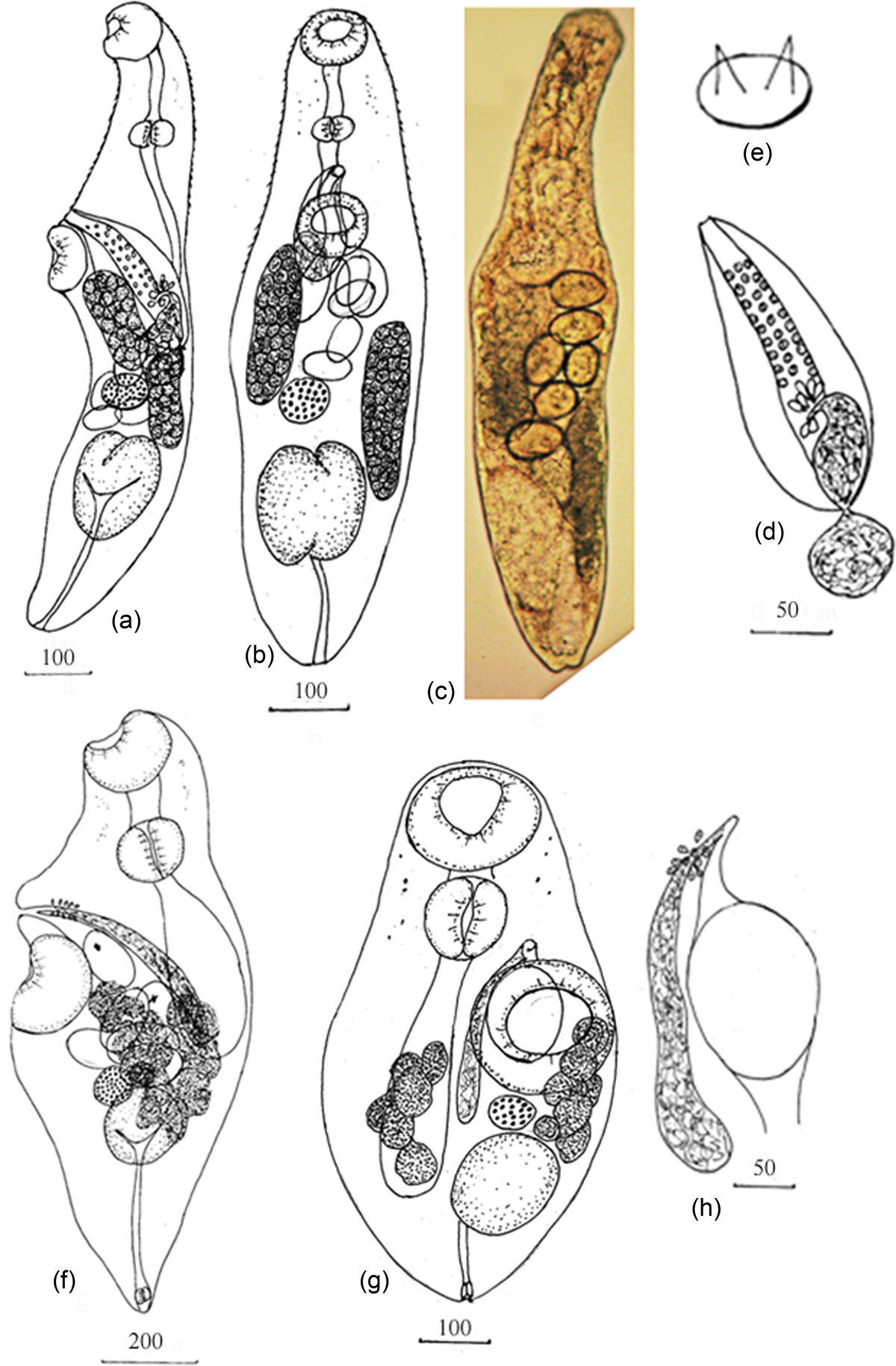
Fig. 1. Parasaccocoelium armatum n. sp.: (a) holotype, lateral view; (b, c) ventral view; (d) hermaphroditic sac; (e) pad with two spines. Pseudohaplosplanchnus catbaensis n. g. n. sp.: (f) holotype, lateral view; (g) ventral view; (h) terminal genitalia. Scale-bars in μm.
Table 3. Measurements (in micrometres) of adult worms of new species.

Molecular data
Two sequences of 28S rDNA fragments 1086 bp in length of P. armatum n. sp. contained no variable sites. The sequences were submitted to the NCBI database with accession numbers MT298950–MT298951.
Remarks
Currently, there are three species within Parasaccocoelium from mullet from south of the Russian Far East (Besprozvannykh et al., Reference Besprozvannykh, Atopkin, Ermolenko and Nikitenko2015). Parasaccocoelium armatum n. sp. is a fourth species that has been found in mugilids from this region. Parasaccocoelium armatum n. sp. is most similar to P. polyovum based on morphology, including the form of the body, testis, ovary, hermaphroditic sac and its reciprocal arrangement and number of eggs within the uterus – more than four (P. mugili, P. haematocheilum have from one to four eggs). However, these species differ from each other by hermaphroditic sac and vitellaria field length and by maximal egg size (table 3). Moreover, vitellaria fields are arranged parallel one to another in P. polyovum and diagonally in P. armatum n. sp. The main difference between P. armatum n. sp. specimens and other Parasaccocoelium species is the presence of an armed hermaphroditic duct. Molecular data support the generic membership of P. armatum n. sp., and, associated with morphological data, indicate a close relationship between the new species and P. polyovum within the monophyletic Parasaccocoelium (fig. 2). Additionally, the genetic p-distance value between P. armatum n. sp. and P. polyovum (0.37% ± 0.19%) is comparable with the interspecific genetic differentiation level for the genus Parasaccocoelium (0.78% ± 0.26%). Nucleotide sequences of 28S rDNA of P. armatum n. sp. and P. polyovum are different by four fixed substitutions.

Fig. 2. Phylogenetic tree of the subfamily Waretrematinae based on the analysis of partial 28S rRNA gene sequences; nodal numbers indicate posterior probabilities for Bayesian inference algorithms.
Family Haplosplanchnidae Poche, 1926
Subfamily Haplosplosplanchninae Poche, 1926
Pseudohaplosplanchnus n. g.
Diagnosis
Body elongated, narrowed posterior end capable of retracting into inside of body. Eyespot pigment dispersed in forebody. Oral sucker subterminal. Prepharynx short. Pharynx transversally oval. Oesophagus absent. Caecum single, reaching level of anterior border ovary. Ventral sucker at level of mid-body, larger than oral sucker. Testis single, in posterior third of body, round or oval. Seminal vesicle tubular, reaching to level posterior border ventral sucker. Pars prostatica thin-walled, surrounded by prostatic cells. Hermaphroditic duct short. Genital pore median, close to anterior border of ventral sucker. Ovary spherical, pre-testicular or contiguous with testis. Seminal receptacle round, contiguous to ovary. Uterus in middle third of body. Eggs large, few, operculated, in distal part of uterus only, containing miracidia with eyespot. Vitellaria in two lateral fields formed from follicles of irregular forms, extending between level of middle ventral sucker and posterior-end testis. Excretory bladder Y-shaped with muscular sphincter. Found in intestine of Mugilidae fishes in Halong Bay, northern Vietnam.
Taxonomic summary
Type species. Pseudohaplosplanchnus catbaensis n. sp.
Etymology. The genus was named Pseudohaplosplanchnus n. g. n. sp. on the basis of morphological similarity of these flukes with representatives of the genus Haplosplanchnus.
Pseudohaplosplanchnus catbaensis n. sp.
Taxonomic summary
Type host. Moolgarda seheli (Forsskål, 1775).
Number of fish examined. 80.
Infection of fish. 5.
Intensity of infection. 1–4 worms per fish.
Site. Intestine.
Type locality. Coastal water of Cat Ba Island, Ha Long Bay, northern Vietnam (20°88′40″N, 10°68′590″E).
Type deposition. Type number 157-Tr, paratype number 158-161-Tr. This material is held in the parasitological collection of the Zoological Museum (Federal Scientific Center of the East Asia Terrestrial Biodiversity Far Eastern Branch of Russian Academy of Sciences, Vladivostok, Russia; e-mail: petrova@ibss.dvo.ru). Deposited: 2019.05.05.
Etymology. Species was named with respect to first description place – Cat Ba Island, Vietnam.
Description
Based on five specimens (fig. 1f–h; table 3). Body elongated, narrowed posterior end capable of retracting into inside of body. Eyespot pigment dispersed in forebody. Oral sucker subterminal. Prepharynx short. Pharynx transversally oval. Oesophagus absent. Caecum single, reaching level of anterior border ovary. Ventral sucker in mid-body larger than oral sucker. Testis single, in posterior third of body, round or oval. Seminal vesicle tubular, reaching to level of posterior border ventral sucker. Prostatic part thin-walled, surrounded by prostatic cells. Hermaphroditic duct short. Genital pore median, close to anterior border of ventral sucker. Ovary spherical, pre-testicular or contiguous with testis. Seminal receptacle round, contiguous to ovary. Uterus in middle third of body, with several loops, reaching level of ovary. Eggs 1–7 in number, large, operculated, in distal part of uterus containing miracidia, with eyespot. Vitellaria in two lateral fields formed from follicles of irregular forms extending, between level of middle ventral sucker and level of posterior-end testis. Excretory bladder Y-shaped with muscular sphincter.
Molecular data
Four successfully amplified and sequenced 18S rDNA of P. catbaensis n. sp. were 1785 bp in length and contained one variable singleton site. Five 28S rDNA fragments 1069 bp in length of P. catbaensis n. g. n. sp. comprised single variable singleton site. The sequences of 18S rDNA and 28S rDNA were submitted to the NCBI database with accession numbers MT298954-MT298957 and MT298959-MT298962, respectively.
Remarks
Pseudohaplosplanchnus catbaensis n. g. n. sp. is similar to representatives of Haplosplosplanchninae by a combination of morphological characteristics, including the presence of a single intestine, single testis and the absence of a cirrus sac. Among haplosplanchnins, these worms are morphologically closer to Haplosplanchnus species. The single difference between Pseudohaplosplanchnus n. g. and Haplosplanchnus is the presence of few (1–7) large eggs (135–142 × 92–104 μm) in the uterus of Pseudohaplosplanchnus n. g. versus numerous small eggs with a maximal size of 75 × 50 μm (Al-Bassel, Reference Al-Bassel1997; Nahhas, Rhodes & Seeto, Reference Madhavi, Jones, Bray and Gibson1997) in the uterus of Haplosplanchnus. Despite the morphological similarity of representatives of these two genera, the validity of Pseudohaplosplanchnus n. g. is supported by molecular data. The 28S rDNA-based genetic distances between Pseudohaplosplanchnus n. g. and Haplosplanchnus are in the intergeneric range, indicating Pseudohaplosplanchnus n. g. does not belong to Haplosplanchnus. On the other hand, molecular data show that the new genus is closely related to Hymenocotta Manter, 1961 (Hymenocottinae Yamaguti, 1971) (fig. 3).

Fig. 3. Phylogenetic tree of the family Haplosplanchnidae based on the analysis of combined 18S rRNA (complete) and of 28S rRNA (partial) gene sequences; nodal numbers indicate posterior probabilities for Bayesian inference algorithms. Sequences from the present study are marked in bold.
Phylogenetic analysis, based on the available molecular data for the Haplosplanchnidae, including type species for Haplosplanchnus and Trigonocephalotrema, revealed that representatives of the Haplosplanchnoidea, Schikhobalotrematinae Skrjabin & Guschanskaja, 1955 and the genus Trigonocephalotrema formed three distinct, highly supported clades. Of these, the Haplosplanchnoidea and Schikhobalotrematinae were closely related to each other with high statistical support, and the genus Trigonocephalotrema appears as a sister clade with poor support (fig. 3). Another highly supported clade contained representatives of the Hymenocottinae, including our new samples of H. mulli and specimens of P. catbaensis n. sp.
The situation with representatives of the genera Trigonocephalotrema and Pseudohaplosplanchnus n. g. is paradoxical. On the one hand, representatives of Trigonocephalotrema possess a combination of morphological characteristics that are representative of worms of the Hymenocottinae and Schikhobalotrematinae (Schikhobalotrema). In particular, specimens of Trigonocephalotrema, like Hymenocotta, possess a peculiar-shaped oral sucker. Pseudohaplosplanchnus n. g. worms are morphologically similar to Haplosplanchnus (Madhavi, Reference Nahhas, Rhodes and Seeto2005). On the other hand, analysis of genetic p-distances and phylogenetic relationships indicate a considerable level of differentiation of Trigonocephalotrema from both Hymenocotta and Schikhobalotrema (p-distance values are 14.2% ± 1.0% and 11.9% ± 0.8%, respectively), and the same for Pseudohaplosplanchnus n. g. and Haplosplanchnus (15.64% ± 1.0%). Overall, p-distance values and the results of phylogenetic analysis of Pseudohaplosplanchnus n. g. (fig. 3) indicate that a new subfamily for this genus, and for Trigonocephalotrema, can be proposed. However, phylogenetic analysis showed that there is a contradiction between the morphological similarity of these worms and their position in the Haplosplanchnidae system, based on the genetic data. Considering these results, we support the view of Huston et al. (Reference Huston, Cutmore and Cribb2018), who state that erecting new subfamilies within the Haplosplanchnidae is questionable because of the lack of molecular data for most haplosplanchnid species; such data are needed to resolve the problematic systematics and phylogeny of this family.
Financial support
The present study was supported by a grant (grant number VAST.ĐA47.12/16-19).
Conflicts of interest
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.








