I. INTRODUCTION
Steroids display a variety of biological functions in the human organism, such as decreasing inflammatory and immune responses (Zeelen, Reference Zeelen1997). Estra-4,9-diene-3,17-dione (Figure 1), a designer steroid, is designed based on the structure of trenbolone (Clarke et al., Reference Clarke, Scarth, Teale, Pearcea and Hillyer2010). This compound shows strong metabolic activation to HuH7 cells (Cooper et al., Reference Cooper, McGrath, Li, Akram, Kasz, Kazlauskas, McLeod, Handelsmanc and Heatherf2017). The major metabolite of this compound is considered to be an isomer of 17-hydroxy-estra-4,9-dien-3-one. Hydroxylation and reduction followed by hydroxylation are the metabolic pathways (Scarth et al., Reference Scarth, Clarke, Teale and Pearce2010). So far, the crystal structure of estra-4,9-diene-3,17-dione has not been reported.

Figure 1. Molecular diagram of estra-4,9-diene-3,17-dione.
II. EXPERIMENTAL
A. Sample preparation
Estra-4,9-diene-3,17-dione (Figure 1) was purchased from J&K Scientific (Beijing, People's Republic of China). The melting point and measured density of estra-4,9-diene-3,17-dione are 145–146 °C and 1.197 g cm−3, respectively. Crystallization of estra-4,9-diene-3,17-dione at room temperature was successful using methanol as a solvent. The crystals are transparent and have a prismatic structure. A portion of the crystals were dried, smashed, screened through 75 μm mesh size, and mounted on a flat zero background plate.
B. Diffraction data collection and reduction
The X-ray powder diffraction measurement was performed at 298 K using an X'Pert PRO diffractometer (PANalytical Co., Ltd., Netherlands) with a PIXcel 1D detector and CuKα radiation (generator setting: 40 kV and 40 mA). The sample was mounted on a flat zero background plate. The diffraction data were collected over the angular range from 4 to 50° 2θ with a step size of 0.01313° 2θ and a counting time of 30 ms step−1.
The software package Material Studio 8.0 (Accelrys Co., Ltd., CA, USA) was used to process the data in the Analytical & Testing Center (Sichuan University, Chengdu, China). The X-ray powder diffraction pattern was pretreated by subtracting the background, smoothing, and stripping off the Kα 2 component. Automatic indexing results were obtained by the X-cell method (Neumann, Reference Neumann2003). The indexing results were then refined using Pawley (R wp = 10.36%) (Pawley, Reference Pawley1981), which involves assigning the Miller indices (hkl) to each observed peak in the experimental PXRD pattern.
C. Single-crystal X-ray diffraction
X-ray diffraction data for estra-4,9-diene-3,17-dione were collected on a New Gemini, Dual, Cu at zero, EosS2 diffractometer. The crystal was kept at 293.15 K during data collection. The structure was solved with Olex2 (Dolomanov et al., Reference Dolomanov, Bourhis, Gildea, Howard and Puschmann2009; Bourhis et al., Reference Bourhis, Dolomanov, Gildea, Howard and Puschmann2015), using charge flipping, and refined with the SHELXL (Sheldrick, Reference Sheldrick2015) refinement package using least-squares minimization.
III. RESULTS
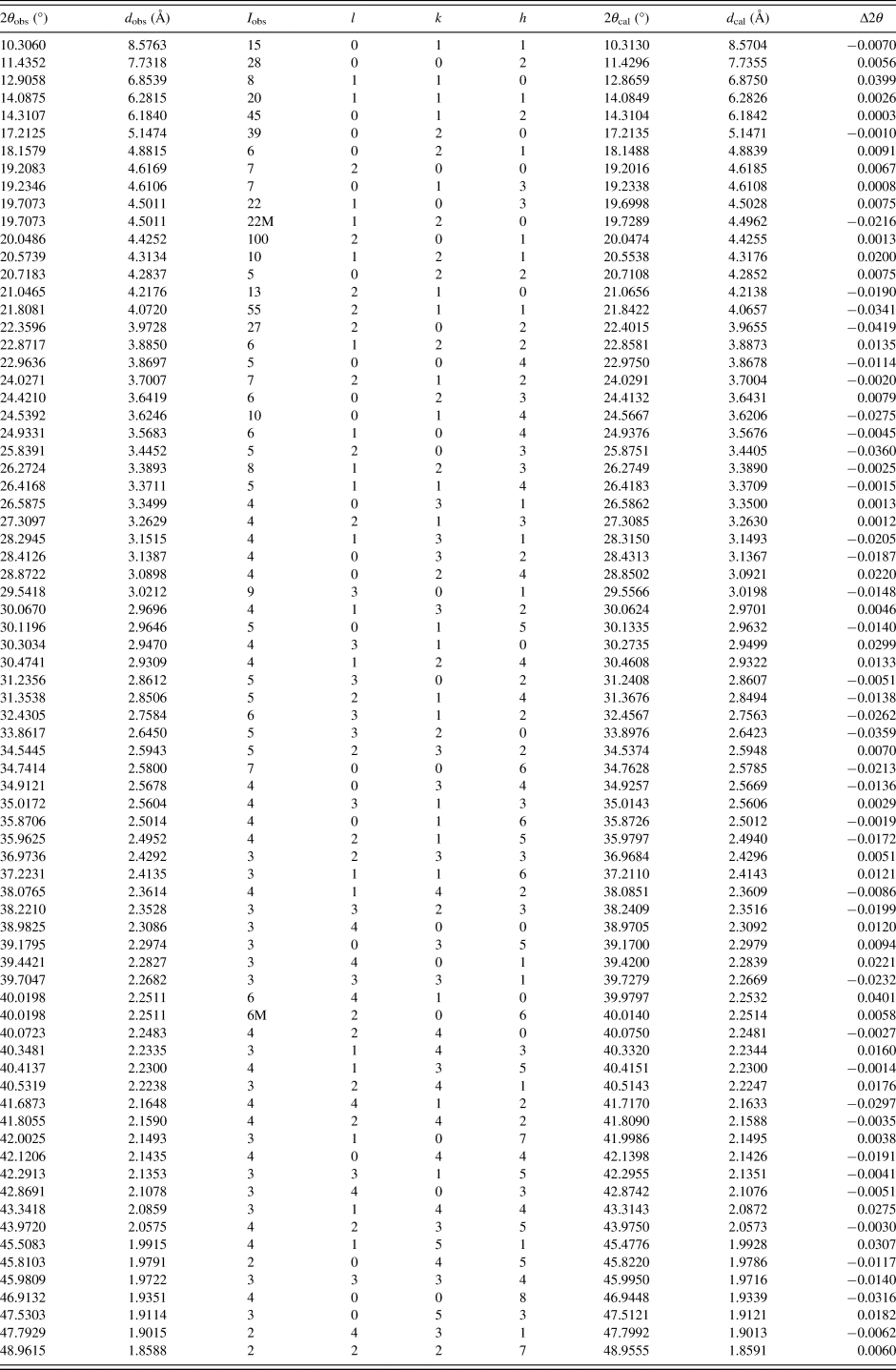
Pawley refinement results confirmed that estra-4,9-diene-3,17-dione is orthorhombic with the space group P212121 and unit cell parameters: a = 9.236(7) Å, b = 10.294(4) Å, c = 15.471(1) Å, unit cell volume V = 1471.11 Å3, Z = 4, ρ cal = 1.221 g cm−3. The values of 2θ obs, d obs, I obs, h, k, l, 2θ cal, d cal, Δ2θ are listed in Table I.
TABLE I. X-ray powder diffraction data for estra-4,9-diene-3,17-dione, C18H22O2. The d-values were calculated using CuKα 1 radiation (λ = 1.54056 Å).
The single-crystal experiment was carried out at the temperature of 293.15 K and the structure solution was obtained [a = 9.2392(7) Å, b = 10.2793(5) Å, c = 15.4822(7) Å, unit cell volume V = 1470.37(15) Å3, Z = 4, ρ cal = 1.221 g cm−3, and space group P212121]. The detailed single-crystal data of estra-4,9-diene-3,17-dione and the experimental data are listed in Table II. The figures were drawn with ORTEP-3 (Oak Ridge Thermal Ellipsoid Plot) and Mercury (Figure 2). Estra-4,9-diene-3,17-dione contains three chiral centers and is arranged in a head-to-tail fashion. The compound is arranged without intramolecular and intermolecular H-bonding, but it has two strong acceptors, namely, C1=O1 and C9=O2.

Figure 2. (a) ORTEP drawing one of the two independent molecules in the asymmetric unit of estra-4,9-diene-3,17-dione with the labeling of non-H atoms. (b) Crystal packing of estra-4,9-diene-3,17-dione.
Table II. Crystal and experimental data of estra-4,9-diene-3,17-dione.

The comparison of PXRD pattern (Deposited Data) with the simulated pattern is shown in Figure 3. Results showed that both single-crystal and powder diffraction methods can get the similar structure data and the deviations of the unit cell parameters and unit cell volume were between 0.03% and 0.15%.

Figure 3. X-ray powder diffraction pattern of estra-4,9-diene-3,17-dione using CuKα radiation (black line) and the simulated pattern of the crystal structure (red line).
IV. DEPOSITED DATA
CIF and/or RAW data files were deposited with ICDD. You may request this data from ICDD at info@icdd.com.
ACKNOWLEDGEMENTS
This work was supported by the Scientific Research Staring Foundation of Yangzhou University, Yangzhou Green Yang Gold Phoenix plans, and Jiangsu Shuangchuang Project.








