Background
Healthcare epidemiology has gained in importance in the United States and Europe due to growing financial pressure on hospitals, rising emergence of multidrug-resistant pathogens and greater complexity of healthcare delivery and systems,Reference Sydnor and Perl 1 , Reference Simmons, Parry, Williams and Weinstein 2 and it is likely to evolve further in the era of big data.Reference Wiens and Shenoy 3
In healthcare, the continuous adoption and integration of electronic medical records, linkage of data sources, and the advent of new diagnostic and digital monitoring technologies have led to an unprecedented quantity and diversity of routine, electronic data.Reference Ross, Wei and Ohno-Machado 4 Big data in healthcare may be used to better exploit the potential for infection prevention and control, quality improvement, and optimal allocation of hospital resources.Reference Wiens and Shenoy 3 , Reference Bates, Saria, Ohno-Machado, Shah and Escobar 5
For healthcare epidemiologists to make use of big data, computational systems and methods that can handle large datasets are required. Parallel with the rising amount of routine healthcare data and improvements in processing speed (computing power doubles every 2 years for the same cost),Reference Moore 6 machine learning is increasingly being used for healthcare projects and is likely to become a key analytical tool in healthcare epidemiology.Reference Wiens and Shenoy 3 , Reference Jordan and Mitchell 7
Thus, digital healthcare epidemiology, which focuses on healthcare populations, may become an important field of epidemiology, analogous to the rapidly growing field of digital epidemiology that uses primarily social media data and other routine data sources within general populations.Reference Salathé 8 – Reference Salathé, Freifeld, Mekaru, Tomasulo and Brownstein 10 Similar to the general field of epidemiology, the primary goals of these interrelated fields, digital epidemiology and digital healthcare epidemiology, are to understand the distribution and determinants of health-related states in specific populations and to use this knowledge to improve health and prevent disease. For simplicity, we characterize the spectrum between conventional healthcare epidemiology and digital healthcare epidemiology across 3 axes: (1) the analytical method, (2) the data source, and (3) the data type (Fig. 1).

Fig. 1 Spectrum between conventional and digital healthcare epidemiology. Note: Any healthcare epidemiology project may be characterized across 3 main axes: the analytical approach, the data source, and the data type as illustrated for a fictive project ‘B.’ This project used nonroutine data from a cohort study and routine data (laboratory and genetic routine data) to predict hospital-acquired infections, primarily via deep learning, a set of machine-learning algorithms, which requires little human guidance for variable selection. Routine healthcare data can be defined as data that are routinely generated or collected during healthcare delivery.Reference Jarow, LaVange and Woodcock 36 Thus, electronic medical records and administrative claims data are typical sources of routine healthcare data.Reference Jarow, LaVange and Woodcock 36 In contrast, nonroutine healthcare data are generated or collected for a specific nonroutine purpose (eg, as part of a clinical trial). Surveillance programs frequently incorporate both routine and nonroutine data sources. Big data is a term used to describe data that make conventional data processing difficult due to their size (volume), diversity (variety), and/or update frequency (velocity).Reference Baro, Degoul, Beuscart and Chazard 37
To exploit the full potential of big routine data in healthcare and to efficiently communicate and collaborate with IT specialists and data analysts, healthcare epidemiologists require some knowledge of large-scale analysis techniques, particularly about machine learning. This review provides an overview on the broad area of machine learning and its recent applications in the emerging field of digital healthcare epidemiology for prediction, detection of trends and patterns (eg, for surveillance purposes), and the identification of risk factors. The main challenges and opportunities of studies relying on routine healthcare data and big data have been reviewed previously.Reference Sips, Bonten and van Mourik 11 – Reference Gray and Thorpe 15
Machine Learning: Introduction
Machine learning as a discipline originated in computer science with very close ties to statistics, but it is difficult to draw a straight line between the two. Machine learning is a young field compared to statistics that arose from the field of mathematics, having developed long before computers became available.Reference Samuel 16 Machine learning and statistics share a common aim to learn from data. Logistic regression for example, which is a standard technique in statistics,Reference Cox 17 is called a machine-learning algorithm within the machine-learning community.Reference Goodfellow, Bengio and Courville 18 The same holds true for more recent algorithms, such as random forests, which are well known machine-learning algorithms, developed by the statistician Leo Breiman.Reference Breiman 19 Most statistical algorithms have been designed to work primarily on small and low-dimensional datasets. The huge and complex datasets that are available today did not exist at the time when the first statistical algorithms were built. The advent of new techniques, and the gathering of huge and complex datasets increasingly required including computational aspects in the algorithms, leading to the term machine learning.
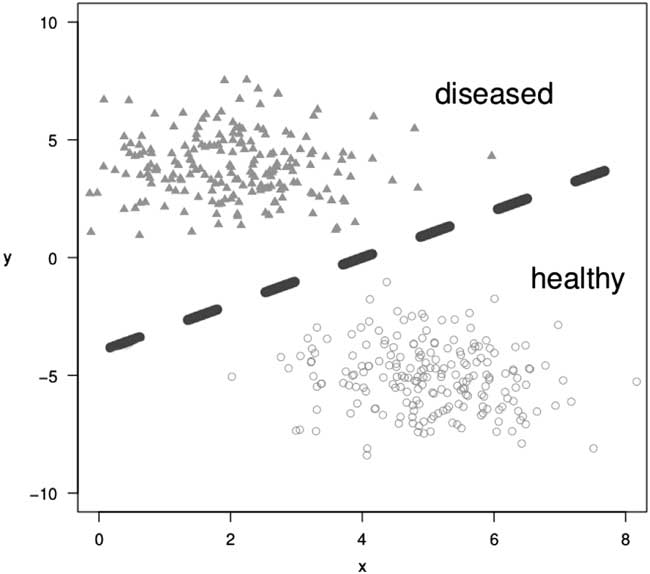
Machine learning can be broadly divided into 2 subareas: supervised and unsupervised learning. In supervised learning, both the input data and the corresponding target values (ie, outcomes) are observed. An example is to classify patients into either diseased or healthy. A domain expert (eg, a physician) assigns an annotation (the “label”), for example, diseased or healthy, to every patient. The aim is to find the model that best distinguishes between those two classes, to either correctly assign the label diseased or healthy to new, unlabeled patients, or to identify important covariables. Problems of this kind are called classification problems (Fig. 2). Well-known classification algorithms include variations of logistic regression, random forests, support vector machines, and neural networks.

Fig. 2 Illustration of a classification task; a model learns to separate diseased from healthy individuals in a 2-dimensional space.
Which classification algorithm is best to use depends on the data type (eg, images, text, laboratory values, and genetic data), as well as the size and the dimensionality of the data. A corresponding model should be carefully selected that will generalize well to unseen data and will not simply memorize the training data (a phenomenon called “overfitting”). Regression algorithms are additional, well-known, supervised machine-learning methods. In regression problems, the aim is not to separate 2 (or more) classes, but to find the function which best describes the data, to predict the correct value for a new data point (Fig. 3).

Fig. 3 Illustration of a regression problem; a model learns the function that best fits the data points.
In unsupervised learning, the training data consists of a set of input variables without any corresponding target values (outcome labels) that are required in supervised learning. The goal in unsupervised learning problems is to find patterns and to extract hidden structure from data, completely data driven without any expert labelling. Typical examples are clustering problems that aim to group similar data points together (Fig. 4). Examples of this type of analysis are subtype detection of patients with hospital-acquired infections and finding similar patient subgroups to assign patients to clinical trials. Typical machine-learning methods used for unsupervised learning are k-means clustering or probabilistic Gaussian mixture models.

Fig. 4 Illustration of an unsupervised clustering task; a model finds similar data points and groups them together.
Machine-learning techniques have become increasingly popular in the last years in the field of healthcare epidemiology due to the huge amount and diversity of routine electronic data that is available in healthcare. However, there still exists a gap between theoretical machine learning research and clinical research. Researchers developing novel machine learning techniques usually have a background in computer science, mathematics or physics. They perform cutting-edge research, develop novel algorithms, and may even apply them to healthcare data. However, they do not have comprehensive knowledge of the data generating process in daily clinical routine. Healthcare professionals, on the other hand, have a deep understanding of the clinical problems and of the quality of specific healthcare data. Today, they are often able to apply some machine learning methods by themselves using standard statistical software packages (eg, R). However, some healthcare professionals may not be aware of the underlying assumptions and limitations of the models, which might lead to statistical unsound models or overfitted models. These 2 research areas complement each other; the advancement of digital healthcare epidemiology to a new level requires mutual understanding, communication, and collaborations between these fields.
Machine Learning: Recent Applications in Digital Healthcare Epidemiology
Numerous recent reports illustrate the first applications of machine learning in digital healthcare epidemiology, most frequently to make predictions based on routine healthcare data (Table 1). This goal is achievable by machine learning, particularly the analysis of large and diverse data assemblages (sometimes involving thousands of variables), which could complicate more human-guided modeling approaches.Reference Jordan and Mitchell 7
Table 1 Recent Applications of Machine Learning in Digital Healthcare Epidemiology

Note. ANN, artificial neural network; CLABSI, central line-associated bloodstream infection; EMR, electronic medical records; I, identification of risk factors; LASSO, least absolute shrinkage and selection operator; NLP, natural language processing; P, prediction; O, other; S, surveillance; SSI, surgical site infection; SVM, support vector machine; XG boost, extreme gradient booster
a Articles published between October 2015 and June 2018 were included based on a Medline search and a bibliographic screening of the selected articles.
b Combines data from different sources (ie, Boston Public Health Commission, Google Trends, Twitter, FluNearYou and electronic medical records).
c Combines data from different sources (ie, Centers for Disease Control and Prevention, electronic medical records, Google Trends, Twitter, FluNearYou, and Google Flu Trends).
In a prototypical, retrospective study based on electronic medical records data from the University of Michigan Hospitals and the Massachusetts General Hospital, Oh et alReference Oh, Makar and Fusco 20 used a data-driven approach to build hospital-specific models to estimate daily patient risk for Clostridioides difficile infection (CDI) using L2 regularized logistic regression. These machine-learning models were built and internally validated based on data from >150,000 adult admissions and involved several thousand time-invariant and time-varying variables; they resulted in a good predictive performance with areas under the receiver operating characteristic curve ranging from 0.75 to 0.82. At both institutions, the models identified half of true-positive cases at least 5 days prior to diagnosis of CDI. As part of a surveillance or decision support tool, these models could help to rapidly identify new cases of CDI, before microbiological test become available, or to select patients who should be tested for the presence of C. difficile. Furthermore, a range of institution-specific predictors were identified (eg, specific departments), which could stimulate additional investigations by health care epidemiologist to generate (causal) hypothesis about risk factors for occurrence or spread of CDI. This study illustrates well the paradigm-shift in healthcare from building ‘one-size-fits-all’ prediction models toward the application of more patient-centered analytical approaches that may result in many different data-driven prediction models. Such a flexible approach can incorporate heterogeneous and changing routine variables, which could complicate the development and application of prediction models that are generalizable across clinics; not all variables can be readily mapped when originating from different systems. Moreover, this approach allows an institution to adjust their models during follow-up, and such flexibility is needed because variables and respective coding practices are subject to change in electronic medical records and because calibration drift may be observed for models derived from regression analysis and machine learning.Reference Davis, Lasko, Chen, Siew and Matheny 21
Even using state-of-the-art machine learning algorithms based on a wide array of potential predictors, some outcomes of interest may not be accurately predicted. This was illustrated recently in a retrospective study by Escobar et alReference Escobar, Baker and Kipnis 22 of granular data from electronic medical records of 21 Kaiser Permanente Northern California hospitals. In this study, none of the conventional and machine learning models discriminated well for prediction of recurrent CDI. Such study results may exemplify that despite the extensive electronic medical records available in this study, relevant predictors may not be readily identified by machine learning or may not even be available in the records.
Furthermore, the external validity and clinical effectiveness of most machine-learning prediction models, like most prediction models and scores in medicine, are unclear. Especially for development and application of predictive models, models should be carefully evaluated in a way that mirrors clinical practice.Reference Sherman, Gurm, Balis, Owens and Wiens 23 Compared to conventional prediction models, machine learning is sometimes a ‘black box’ approach, such that selection of predictor variables by machine-learning algorithms may not be transparent and can be counterintuitive.Reference Oh, Makar and Fusco 20 However, predictive modeling via machine learning (like other statistical techniques) does not require including only causal predictors, because accurate prediction models can be derived from an abundance of variables and proxy measures of causal factors that may not be causally related to the outcome of interest (eg, brain natriuretic peptide being a marker for heart failure).
Compared to prediction tasks, little has been reported about using machine learning in healthcare epidemiology to draw causal inferences and to identify independent risk factors (ie, causal modeling), which requires careful consideration of bias, confounding, interactions, reverse causality, and effects by chance.Reference Neugebauer, Schmittdiel and van der Laan 24 Some machine-learning algorithms can detect linear and nonlinear interactions between variables, but confounding may be challenging to address adequately without human guidance through conceptual causal frameworks and expert knowledge. Implementation of machine learning algorithms that can account for observed confounding have been proposed and may become more advanced in the future.Reference Lippert, Casale, Rakitsch and Stegle 25 – Reference Li, Rakitsch and Borgwardt 27 Thus, causal hypotheses could be generated from routine healthcare data via machine learning with little human guidance (eg, to identify a hidden outbreak source). However, application of machine learning in observational studies cannot replace adequately sized and well-executed randomized controlled trials in making causal inferences because randomized controlled trials account for both known and unknown/unmeasured confounding.
Most recent studies applying machine learning to real-world tasks in healthcare epidemiology relied, at least partly, on routine data originating from electronic medical records (Table 1). Such rich data sources have been shown to be especially useful for developing hypotheses about previously unknown risk factors and for building accurate prediction models for various outcome of interest (eg, specific healthcare-acquired infections, hospital complications).Reference Oh, Makar and Fusco 20 , Reference Beeler, Dbeibo and Kelley 28 – Reference Savin, Ershova and Kurdyumova 30
Linkage of electronic medical records data with high-quality cohort or registry data has become a valuable option to add exposures, potential confounders, effect modifiers, and outcomes of interest with strict definition criteria that may not be present in routine medical records.Reference Cook and Collins 31 This option is particularly valuable because data from electronic medical records (and other routine data sources) have been reported to sometimes be of lower quality than data acquired during prospective investigations due to changing and varying definition criteria/coding practices, and missing data.Reference Cook and Collins 31 , Reference Benchimol, Smeeth and Guttmann 32 Therefore, studies relying on routinely collected health data require careful consideration of potentials for information bias, selection bias, and residual confounding at the design stage and analytical stage of the study. Furthermore, reporting of respective study results should be as transparent as possible.Reference Benchimol, Smeeth and Guttmann 32
In addition to structured, routine data elements, unstructured data (eg, clinical notes) can now provide reasonable information when analyzed by the machine-learning method of natural language processing; this approach can further increase the volume of accessible, routine healthcare data.Reference Ford, Carroll, Smith, Scott and Cassell 33 However, the incremental value of healthcare data obtained from daily routine clinical notes is not proven, and both unstructured and structured routine data may not always be reliable and suitable.Reference Sips, Bonten and van Mourik 11
To utilize the increasing volumes of routine healthcare data from health records and other routine data sources, concerns about data quality, data heterogeneity, missing data, and selective data collection are important to consider for any machine learning task; the main challenges and opportunities of studies relying on routine healthcare data and big data have been reviewed previously.Reference Sips, Bonten and van Mourik 11 – Reference Gray and Thorpe 15
Gaps in Knowledge
To date, little has been reported about applications of state-of-the-art machine learning to healthcare epidemiology. Specifically, the efficacy and effectiveness of machine-learning–derived prediction models to improve healthcare delivery has yet to be proven.Reference Wiens and Shenoy 3 Notably, it remains largely unknown how machine learning could be adequately translated into clinical practice. Therefore, more research is required to elucidate the good, the bad, and the unintended consequences of machine learning in healthcare epidemiology and to understand how to best apply machine learning findings to healthcare practice.Reference Cabitza, Rasoini and Gensini 34 Despite many sensational media reports, machine learning it not a magic technology that can convert data of poor quality into goldReference Beam and Kohane 35 and, as a data scientist has stated recently, “Machine learning in healthcare is still the wild west.”
The increasing volume, variety and velocity of routine healthcare data clearly provide massive potential for supervised and potentially unsupervised machine learning tasks in healthcare epidemiology. However; to make optimal use of (big) routine data for quality improvement and healthcare research, these developments should be met by appropriate methodological, ethical, and data security standards.
In conclusion, digital healthcare epidemiology is a growing field in medicine that is driven by the increasing availability of big data originating from daily routine documentation in healthcare. Machine learning may become an important tool in the armamentarium of healthcare epidemiologists to better exploit the potential of big data for infection prevention and control, quality improvement, and optimal allocation of hospital resources. Due to their complexity, machine-learning projects should usually be performed in close collaboration between domain experts and machine-learning specialists based on best practices.
Acknowledgments
Financial support
This work was funded by the Division of Infectious Diseases and Hospital Epidemiology, University Hospital Basel, Basel, Switzerland.
Conflicts of interest
All authors report no conflicts of interest relevant to this article.