Introduction
Arctic ground squirrels (Urocitellus parryii; synonym: Spermophilus parryii) were common small mammals of the late Pleistocene fauna in Beringia (Harington, Reference Guthrie2011), the vast unglaciated region between the Kolyma River (alternatively the Lena River) in eastern Siberia and the Mackenzie River in the Northwest Territories (Canada), including the intervening continental shelf shallower than ca. 200 m below current sea level (Matthews, Reference Mann, Peteet, Reanier and Kunz1982; Elias and Brigham-Grette, Reference Edwards and Armbruster2007). Arctic ground squirrels, along with most small to medium sized mammals, survived the major extinction event at the end of the Pleistocene and still live across the Holarctic region (although not in the study area). Male arctic ground squirrels tend to inhabit dry, well-drained sites and construct underground rooms or hibernacula roughly 1 m below ground in the permafrost active layer, and line their nests with grasses. They cache large quantities of fruits and seeds in the hibernacula alongside their nests, and feed on the cached plants in early spring. This allows the ground squirrel to regain body weight that was lost during hibernation, and to be more successful at competing and mating directly upon emergence from their burrows (Buck and Barnes, Reference Buck and Barnes1999; Zazula et al., Reference Zazula, Mathewes and Harestad2006a).
Arctic ground squirrel nests and caches, together called middens, found preserved in Pleistocene aged permafrost deposits across Beringia contain highly detailed records of former local vegetation, as was shown through recent macrofossil studies on Beringian specimens (Zazula et al., Reference Zazula, Froese, Elias, Kuzmina and Mathewes2005, Reference Zazula, Froese, Elias, Kuzmina, La Farge, Reyes, Sanborn, Schweger, Smith and Mathewes2007, Reference Zazula, Froese, Elias, Kuzmina and Mathewes2011; Gaglioti et al., Reference Froese, Zazula and Reyes2011). As arctic ground squirrels are selective in the species they collect (Zazula et al., Reference Zazula, Mathewes and Harestad2006a), using only macrofossil analyses may generate an incomplete or biased view of Pleistocene vegetation (Zazula et al., Reference Zazula, Froese, Westgate, La Farge and Mathewes2005). Microfossil analyses, including pollen, non-pollen palynomorphs (NPPs), and phytoliths, can begin to fill gaps in vegetation reconstructions resulting from the ground squirrel’s selectivity bias of plant material. Charcoal recovered from ground squirrel nests also provides information as to whether fire was present in the landscape at the time of nest construction. Ancient DNA (aDNA) has also proven to be an important source of information about ancient plants, fungi and animals (Willerslev et al., Reference Willerslev2014).
A combined approach of macrofossil, microfossil, and aDNA analyses has yet to be performed on these middens, or on most paleoecological archives from the area. Here, we provide a multi-proxy approach using macrofossils, microfossils, and aDNA to reconstruct a time-slice of glacial history in the Yukon Territory. We discuss the implications of using a multi-proxy approach to improve local and regional vegetation reconstructions in depositional environments such as middens, by removing the selectivity bias created by ground squirrels to a significant extent.
Material and methods
Materials
The midden was located at a small placer goldmine on Little Blanche Creek, Klondike area near Dawson City, Yukon Territory, Canada (63°50'35''N 139°05'41''W) in July 2015. Dawson tephra, a well-known stratigraphic marker that dates to ca. 25,300 14C yr BP (Westgate et al., Reference Westgate, Preece, Kotler and Hall2000; Froese et al., 2006) that is commonly exposed in the permafrost deposits at many of the Klondike placer gold mines, was observed in the profile some 1.5 m above the midden. Skeletal elements of Wisconsinan (Rancholabrean Land Mammal Age, Weichselian in Europe, last glacial period) vertebrates, like steppe bison (Bison priscus), horse (Equus spp.), reindeer (Rangifer tarandus), and woolly mammoth (Mammuthus primigenius) were commonly recovered at the site. Some arctic ground squirrel middens (identified based on their large size) were noticed in the thawing exposure of the mine, though most were inaccessible for study. One almost complete midden that was accessible from the valley bottom was collected (Fig. 1), kept frozen in a standard cooler box with ice packs and transported back to the field camp and air dried. Material collected consisted of one almost complete arctic ground squirrel midden (volume ca. 1 L) and some small lumps (<1 cm3) of surrounding sediment.

Figure 1 (color online) The midden of an arctic ground squirrel Urocitellus parryii we collected in-situ at the Little Blanche Creek in Yukon, Canada.
Subsampling
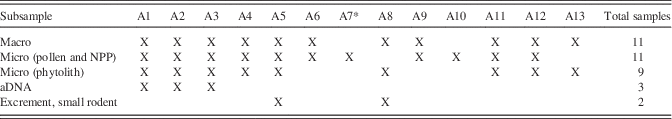
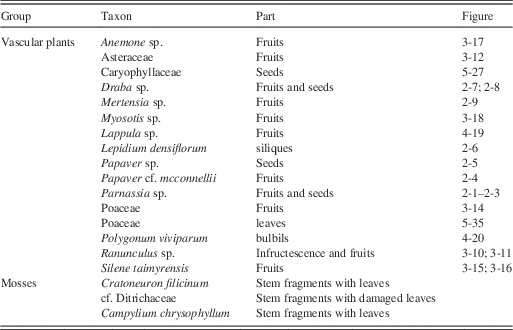
All visually different constituents in the midden were sampled, e.g., coherent blocks of ground squirrel midden of different structures (such as grass spikelets, fibrous tissue of varying size, and non-poaceous material). Each sample was then subsampled for laboratory processing of the individual proxies to be included in the analysis (i.e., macrofossils, pollen, phytoliths, charcoal, and aDNA; Table 1). Subsamples for aDNA were taken from the inner-most core of coherent blocks of vegetative material; subsampling was performed in the aDNA laboratory of Naturalis Biodiversity Center using standardized protocols to prevent contamination (Cooper and Poinar, Reference Cooper and Poinar2000). Subsamples for macrofossils and microfossils were obtained and processed at the University of Amsterdam Paleoecology Laboratory. Fecal pellets that were encountered in macrofossil samples were also sampled for pollen content.
Table 1 Overview of available subsamples and proxies studied.

*This subsample consisted of clay (surrounding sediment), while all others were vegetative material from the midden.
Radiocarbon dating
One subsample of poaceous (grass) material from the Little Blanche Creek midden was submitted to the Centre for Isotope Research (CIO) at the University of Groningen (The Netherlands) for AMS radiocarbon dating.
Macrofossils
Eleven macrofossil subsamples were analyzed (Table 1). Subsample volumes for macrofossils were estimated by volumetric displacement of 5% KOH solution. Macrofossils were retrieved after warming the subsamples in 5% KOH under regular careful stirring with a glass rod to enhance disaggregation, followed by washing over a 160 μm mesh sieve. The remaining material was suspended, small volumes at a time, in water on a petri dish, and examined systematically using a Leica MZ16 stereomicroscope at ca. 12.5× magnification (Birks, Reference Birks2007; Mauquoy et al., Reference Mauquoy and van Geel2011). Fruits, seeds, selected vegetative remains, and non-plant macrofossils were individually picked and identified. Vascular plant specimens were identified using the literature (Lawton, Reference Kołaczek, Zubek, Błaszkowski, Mleczko and Margielewski1971; Crum and Anderson, Reference Crum and Anderson1981; Vitt and Buck, Reference Vitt and Buck2001; Zazula et al., Reference Zazula, Froese, Elias, Kuzmina and Mathewes2005, Reference Wooller, Zazula, Blinnikov, Gaglioti, Bigelow, Sandorn, Kuzmina and La Farge2006b, Reference Zazula, Mathewes and Harestad2006c, Reference Zazula, Froese, Elias, Kuzmina, La Farge, Reyes, Sanborn, Schweger, Smith and Mathewes2007, Reference Zazula, Froese, Elias, Kuzmina and Mathewes2011; Cappers et al., Reference Cappers, Bekker and Jans2006; Mauquoy and van Geel, Reference Matthews2007; van Geel et al., Reference van Geel and Aptroot2008, Reference van Geel, Fisher, Rountrey, van Arkel, Duivenvoorden, Nieman, van Reenen, Tikhonov, Buigues and Gravendeel2011a, Reference van Geel, Buurman, Brinkkemper, Schelvis, Aptroot, van Reenen and Hakbijl2011b; Gaglioti et al., Reference Froese, Zazula and Reyes2011; Wooller et al., Reference Wooller, Zazula, Blinnikov, Gaglioti, Bigelow, Sandorn, Kuzmina and La Farge2011; The Plant List, Reference Taberlet, Coissac, Pompanon, Gielly, Miquel, Valentini, Vermat, Corthier, Brochmann and Willerslev2013; Gravendeel et al., Reference Goecks, Nekrutenko and Taylor2014), the reference collection housed in the Institute for Biodiversity and Ecosystem Dynamics (IBED) at the University of Amsterdam, and the expert opinions of specialists. Selected macrofossils were deposited in the collections at the Natural History Museum Rotterdam, The Netherlands (collection numbers NMR 999700003521, NMR 999900011869, NMR 999900011880 through NMR 999900011888). The remainder of the sampled midden was also deposited there for future analyses (NMR 999900011871).
Microfossils
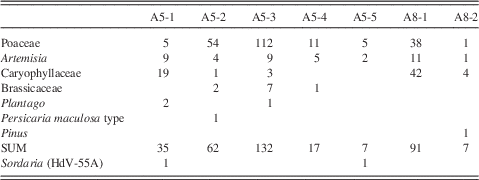
Eleven samples from the Little Blanche Creek midden were analyzed for pollen and non-pollen palynomorphs (NPPs, mainly fungal spores; Table 1). Of the 11 samples, 10 were collected from nest material in the midden and 1 (sample A7) consisted of sediment from near the midden. Two subsamples (A5 and A8) contained small (ca. 8×3 mm) cylindrical fecal pellets. At that size, they are too small to have been produced by an arctic ground squirrel, but likely originated from a microtine rodent (Zazula et al., Reference Zazula, Froese, Westgate, La Farge and Mathewes2005). A total of 5 subsamples from the fecal pellets in the A5 sample and 2 subsamples from the fecal pellets in the A8 sample were also analyzed for pollen content (Table 1).
The preparation of all pollen and NPP samples followed Chambers et al. (Reference Chambers, van Geel and van der Linden2011: table 2). Slides were counted using a Leica DMLB stereomicroscope at 400× magnification, with pollen sums that exceeded 300 pollen grains of terrestrial taxa. Percentages of reworked pollen, Cyperaceae, fern spores, and NPP were expressed on the terrestrial pollen sum (Chambers et al., Reference Chambers, van Geel and van der Linden2011). In samples with one very abundant dominant taxon, extended scans were performed and the occurrence of any additional pollen types was noted (+). Pollen grains and spores were identified using the IBED reference collection, the literature (McAndrews et al., Reference Mauquoy, Hughes and van Geel1973; van Geel, 1978; Kapp et al., Reference Juggins2000; Beug, Reference Beug2004; van Geel and Aptroot, Reference van Geel2006) and expert opinions of specialists. NPP type numbering and laboratory codes follow Miola (Reference McAndrews, Berti and Norris2012). All microfossil slides were deposited in the collection of the Natural History Museum Rotterdam (NMR 999900011870) and residues are kept in the collection of IBED (numbers G 30.306 through G 30.315, G 30.539 through G 30.545, and G 30.387).
Phytoliths, silica-based remnants of vegetative plant material, were extracted from nine subsamples of midden material and prepared according to standard laboratory procedures (Piperno, Reference Morcote-Ríos, Giraldo-Cañas and Raz2006). At least 200 phytoliths per sample were counted under a Zeiss Axiophot Photomicroscope at 400× magnification with Differential Interference Contrast (DIC) for enhanced viewing of silica, which has the same refractive index as glass. Phytolith morphotypes were identified using the reference collection housed at IBED at the University of Amsterdam, reference atlases (Piperno, Reference Morcote-Ríos, Giraldo-Cañas and Raz2006) and expert opinions.
We used multivariate ordinations to examine the similarity and dissimilarity of pollen and phytolith assemblages. Pollen, NPP, and phytolith data were analysed using detrended correspondence analysis (DCA). The results from DCA are also in terms of standard deviations, which allowed for comparisons of outputs between proxies. All analyses were performed in RStudio Version 1.0.136 (R Development Core Team, Reference Pirozynski, Carter and Day2011). Pollen and phytolith diagrams were plotted with C2 software (Juggins, Reference Jacobson and Bradshaw2003).
aDNA
The arctic ground squirrel midden was loosely disaggregated and air-dried before shipping in a closed bag. During subsampling, care was taken to avoid any possible contamination: only material that could be taken from inside compact lumps of vegetation from the arctic ground squirrel midden was used. All pre-PCR (Polymerase Chain Reaction) work, including subsampling, was done in the aDNA lab of Naturalis Biodiversity Center, The Netherlands. No work on ground squirrels was previously performed in this lab and all extractions and PCRs included blanks (Cooper and Poinar, Reference Cooper and Poinar2000).
Silica extraction (based on Rohland and Hofreiter, Reference Richardson and Watling2007) was used to obtain aDNA. Subsamples (ca. 1 mL) were ground in a Retsch CryoMill at –196°C (set to: automatic precooling followed by two cycles [40 s each] of grinding at 30 Hz). Powdered subsamples were suspended in silica extraction buffer and incubated overnight in a hybridizer rotor in the dark at room temperature (RT). After centrifuging, the supernatant was transferred into an L2 buffer with silica suspension and the pH was adjusted to ca. 4.0 by adding small volumes of HCl. After 3h incubation (hybridizer rotor, RT, in the dark), extracts were purified by repeated centrifugation and addition of fresh buffer, finally using a New Wash buffer. Then extracts were centrifuged, the supernatant was discarded and pellets were air-dried, finally to be re-suspended in 50 µL of TE and stored at –20°C.
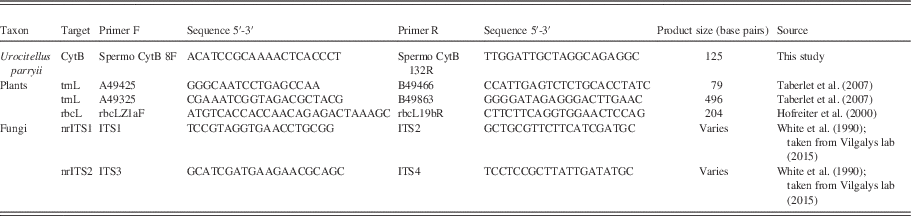
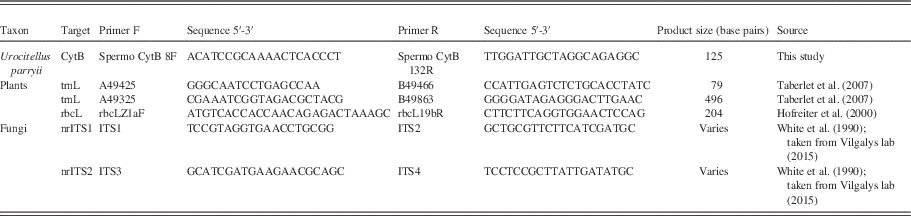
Standard barcode markers were selected to amplify specific taxa, i.e., animals, fungi and plants (i.e., any organism with chloroplasts with chlorophyll b; Cavalier-Smith, Reference Cavalier-Smith1981), including the mitochondrial CytB gene for the arctic ground squirrel, the chloroplast genes trnL (Taberlet et al., Reference Stalpers2007) and rbcL (Hasebe et al., Reference Harington1994) for plants, and nuclear ribosomal genes nrITS1 and nrITS2 for fungi (Schoch et al., Reference Rohland and Hofreiter2012; Table 2). Our methods follow van Geel et al. (Reference van Geel, Fisher, Rountrey, van Arkel, Duivenvoorden, Nieman, van Reenen, Tikhonov, Buigues and Gravendeel2011a) for plants and fungi. Novel primers were designed in Geneious 9.0.4 (http://www.geneious.com; Kearse et al., Reference Kapp, Davis and King2012) using modern Urocitellus parryii sequences from NCBI GenBank. All primers included M13-tails. The arctic ground squirrel primers were used to confirm the identity of the species that built the midden (Hofreiter et al., Reference Hasebe, Omori, Nakazawa, Sano, Kato and Iwatsuki2000).
Table 2 Primers used in this study. Product size indicates the total size of the obtained products, including the primers.
aDNA extracts were diluted 1:5. PCRs of ancient material were carried out on a Bio-Rad C1000 Touch Thermal Cycler or Bio-Rad S1000 Thermal Cycler using Phire Hot Start DNA Polymerase. The PCR consisted of a 30 s activation step at 98°C, followed by 35–40 cycles at 98°C for 5 s, 50°C for 5 s and 72°C for 10 s, with a concluding step at 72 °C for 1 min. After initial PCRs produced amplicons, they were rerun using the same PCR schedule with MID-labelled primers (trnL, rbcL, and nrITS) for Ion Torrent sequencing. The amplicons from the arctic ground squirrel primers were Sanger sequenced by BaseClear B.V. (Leiden, The Netherlands) on an ABI3730XL sequencer (Life Technologies) to check their specificity.
Resulting PCR products were run on a QIAxcel machine to measure sizes and concentrations. Equimolar pools were made based on these results. One pool contained all of the small fragments (trnL and rbcL) and fragments generated with A49325+B49863 (trnL), which contained mainly small products. The other pool contained products from both nrITS regions. Using Ampure XP beads from Agencour, size selection was performed and primer dimers were removed from the generated PCR pools. The beads were washed with 150 mL 70% EtOH twice and resuspended in 40 µL MilliQ water. Cleaned PCR products were quantified using an Agilent 2100 Bioanalyzer DNA High sensitivity chip. The equimolar pools were diluted according to the calculated template dilution factor to target 10–30 % of all positive Ion Sphere Particles. Template preparation and enrichment was carried out with the Ion PGM™ Template OT2 200 (small fragments) and Ion PGM™ Template OT2 400 (large fragments) kits with use of the Ion One Touch System, according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA). Quality control of the Ion One Touch 200 Ion Sphere Particles was done with the Ion Sphere Quality Control Kit using a Life Qubit 2.0. The pool containing the small fragments held 32.4% templated ISPs measured with the Qubit and the pool with the large fragments firstly held 82% templated ISPs. After further dilution, the last pool contained 11.2% templated ISPs. The Enriched Ion Spheres were prepared for sequencing on a Personal Genome Machine (PGM) with the Ion PGM™ Hi-Q™ Sequencing Kit according to the manufacturer’s protocol (Life Technologies) and deposited on an Ion-314-chip (520 cycles per run) in a single loading cycle for one sequencing run.
Sequences obtained (Supplementary Tables 1–6) were analyzed in Galaxy (Giardine et al., Reference Geml, Kauff, Laursen and Taylor2005; Blankenberg et al., Reference Blankenberg, von Kuster, Coraor, Ananda, Lazarus, Mangan, Nekrutenko and Taylor2010; Goecks et al., Reference Gill, McLauchlan, Skibbe, Goring, Zirbel and Williams2010). They were sorted, clustered using CD HIT, and serially BLASTed (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990) against NCBI GenBank (trnL and rbcL products) and the UNITE Fungal ITS Database (ITS products) using the BLAST Wrapper for online or local BLASTing integrated in Galaxy (Camacho et al., Reference Camacho, Coulouris, Avagyan, Ma, Papadopoulos, Bealer and Madden2009). Only matches above 50 bp long (70 bp for the nrITS fragments) with a minimal mean quality of 20 or higher and at 97% or more similarity to reference data were used. To compensate for any incompleteness of the reference databases and short reads, resulting identifications were manually culled to genus level maximally. These were interpreted using the literature (Richardson and Watling, Reference Reimer, Bard, Bayliss, Beck, Blackwell, Bronk and Buck1997; Cody, Reference Cody2000; Flora of North America, Reference Elias2016; Vascular Plants of Canada (VASCAN) Database, 2016) and screened for aberrant records, such as tropical plants. In some samples, a few (<10) of these records were found, standardly present in aDNA sequencing projects such as Cannabis and Emmotum. As the corresponding blanks were clean, these could not have been caused by contamination in the lab. If contamination had occurred, it was between collecting in the field and entry into the aDNA lab. However, these records could also have resulted from sequencing errors, errors in the reference databases, or non-diagnostic reads. These non-informative sequences were excluded from further interpretation.
Results
Radiocarbon dating
The selected poaceous (grass) macrofossils from the Little Blanche Creek midden were radiocarbon dated to 26,210 ± 140 14C yr BP (GrA-64905), which calibrates to 30,740–30,380 cal BP using IntCal13 (Reimer et al., 2013). This date is in stratigraphic alignment with the younger overlaying tephra marker bed, which was deposited ca. 27,000 cal yr BP.
Macrofossils
The macrofossil samples (n=11) yielded a combined volume of 79 mL, consisting of 13 genera (Table 3, Fig. 2–5). Poaceous remains consisted of fruits and spikelets (Fig. 3-13 and 3-14) and vegetative remains. Most macrofossils samples were, however, dominated by Poaceae (Table 4). Overall, ca. 84% of 79 mL consisted of Poaceous remains, while the other 16% consisted of a variety of non-Poaceous plant remains. The non-poaceous macrofossil assemblages were dominated by Draba fruits (Fig. 2-7) and seeds (Fig. 2-8), Lepidium densiflorum siliques (Fig. 2-6), and Papaver, Ranunculus and Parnassia fruits and seeds (Fig. 2-1–2-5 and Fig. 3-10 and 3-11). All other taxa (Table 3) occurred in lower abundances. The mosses Campylium chrysophyllum and Crataneuron filicinum each occurred in only one subsample. Fungal fruit-bodies were encountered in one subsample, which consisted of fibrous tissue with many hairs (Fig. 5-37). The majority of the hairs were short and formed a quite compact wall from which many long hairs emerged, which ended in hooks, with which the fruit-bodies interlocked. These fruit-bodies were tentatively identified as Chaetomium sp. using Ames (Reference Ames1961). Compared to Ames’ (Reference Ames1961) illustrations, the long hairs are relatively short, but the overall habitus fits very well. After photography, the fruit-bodies were squashed on a microscope slide, but no spores were obtained.

Figure 2 (color online) Vascular plant remains from the arctic ground squirrel midden. (1) Parnassia fruit with seeds. (2) Parnassia seeds. (3) Parnassia seed (left) and seed removed from envelope. (4) Papaver cf. mcconnellii fruit. (5) Papaver seeds. (6) Lepidium densiflorum siliques. (7) Draba fruits. (8) Draba seeds. (9) Mertensia fruits.

Figure 3 (color online) Vascular plant remains from the arctic ground squirrel midden. (10) Ranunculus infructescence. (11) Ranunculus fruits. (12) Asteraceae fruit. (13) Poaceae spikelets. (14) Poaceae fruits. (15) Silene taimyrensis fruit. (16) Silene taimyrensis seeds. (17) Anemone fruits. (18) Myosotis fruit.

Figure 4 (color online) Vascular plant remains from the arctic ground squirrel midden. (19) Lappula fruit. (20) Polygonum viviparum bulbil. (21) Unidentified fruit and seeds. (22) Unidentified fruit and seeds. (23) Unidentified seed. (24) Unidentified fruits and seeds. (25) Unidentified fruit and seeds.

Figure 5 (color online) Vascular plant (including phytoliths), bryophyte, animal and fungal remains from the arctic ground squirrel midden. (26) Unidentified fruit and seeds. (27) Caryophyllaceae seed. (28) Unidentified fruit and seed. (29) Unidentified fruit and seeds. (30) Unidentified seed. (31) Unidentified seed. (32) Unidentified “seed.” (33) Fecal pellet of microtine rodent. (34) Unidentified bryophyte sporangium. (35) Unidentified fungal fruit-bodies on poaceous tissue. (36) New fungal ascospore type HdV-834. (37) Chaetomium fruit-bodies. (38) Unknown morphotype A phytolith. (39a) Top view of wavy trapezoid phytolith. (39b) Side view of wavy trapezoid phytolith. (40a) Side view of rondel phytolith. (40b) Top view of rondel phytolith. (41) Tall saddle phytolith.
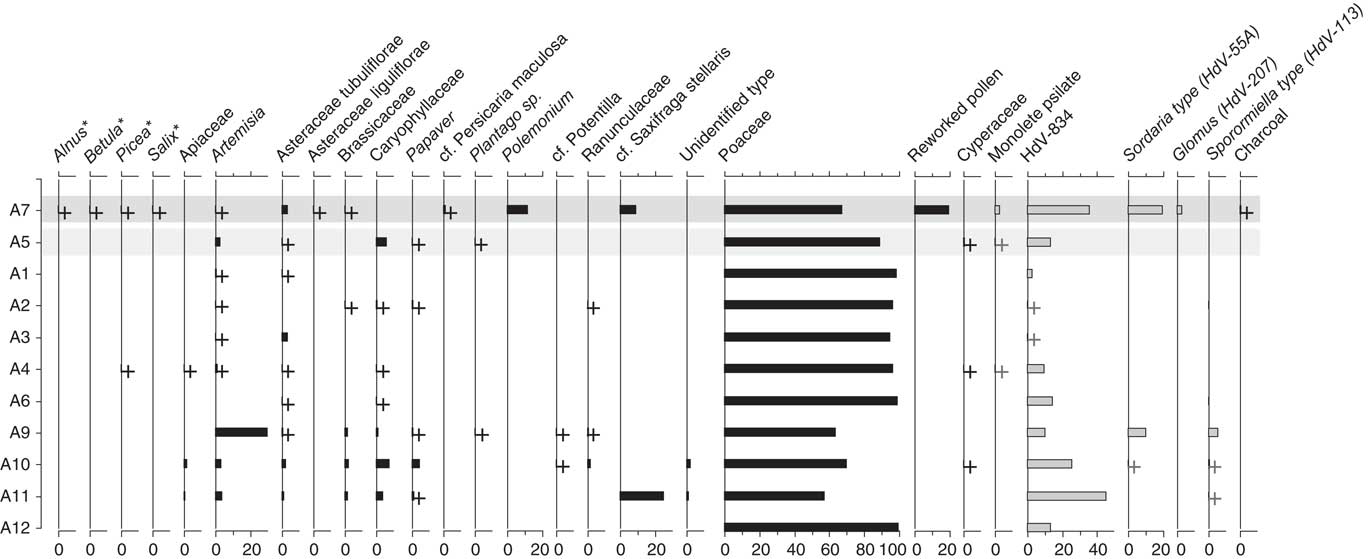
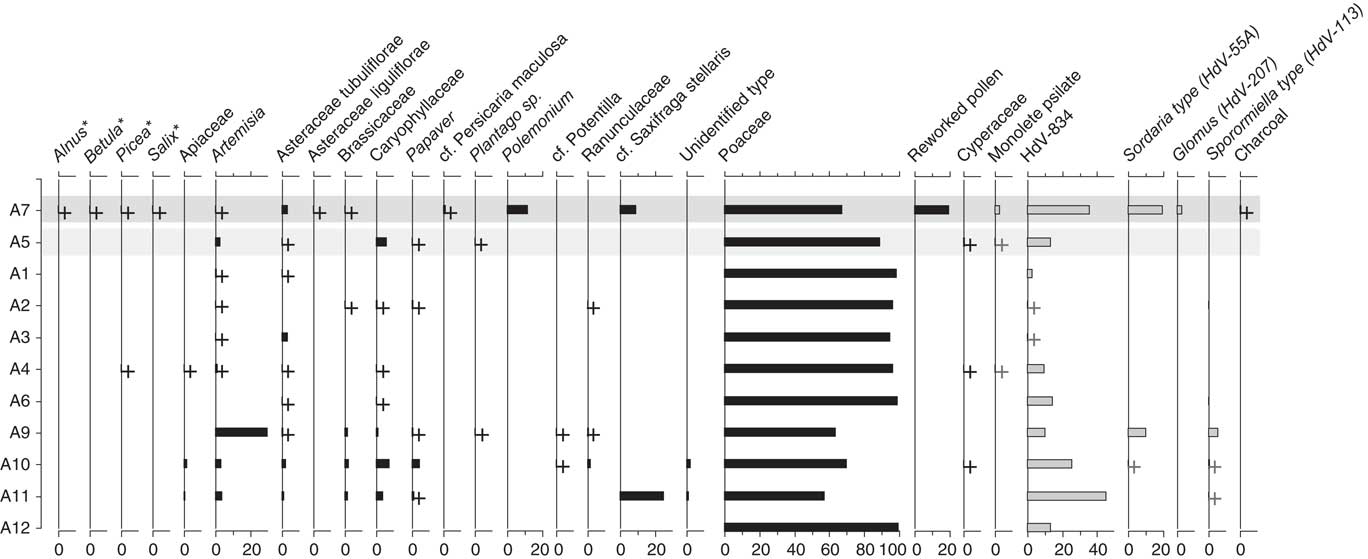
Figure 6 Pollen and non-pollen palynomorph (spore) assemblages from the Little Blanche Creek midden nest material and nearby sediment. Dark gray shading across the sample represents the sediment sample (A7) and the light gray shading represents nest material samples containing microtine fecal pellets (A5). Samples are listed on the y-axis, and the x-axis is in units of percentage. Plus signs represent presence of a given taxon in a sample, though the percentage was too low to visualize. Black bars represent pollen types and gray bars with black outlines indicate spore types. Arboreal genera are marked with an asterisk (*).

Figure 7 (color online) Artemisia pollen grain from sediment subsample A7 interpreted as having been reworked.
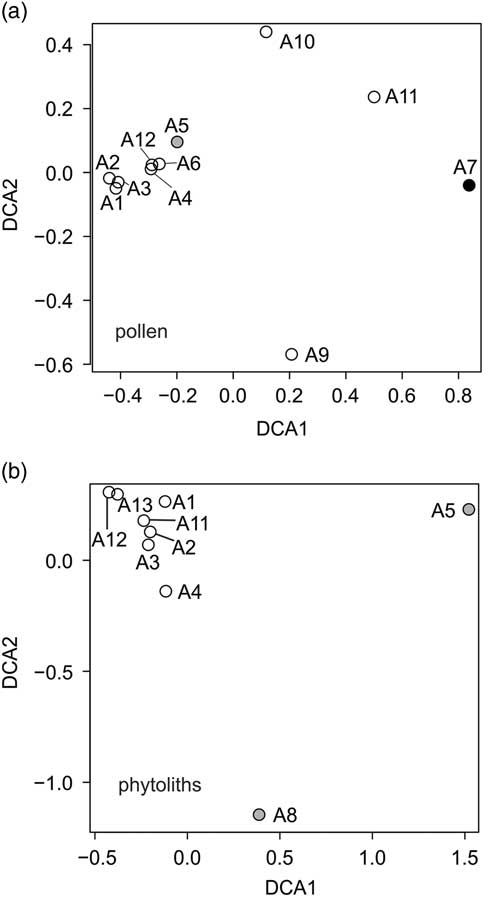
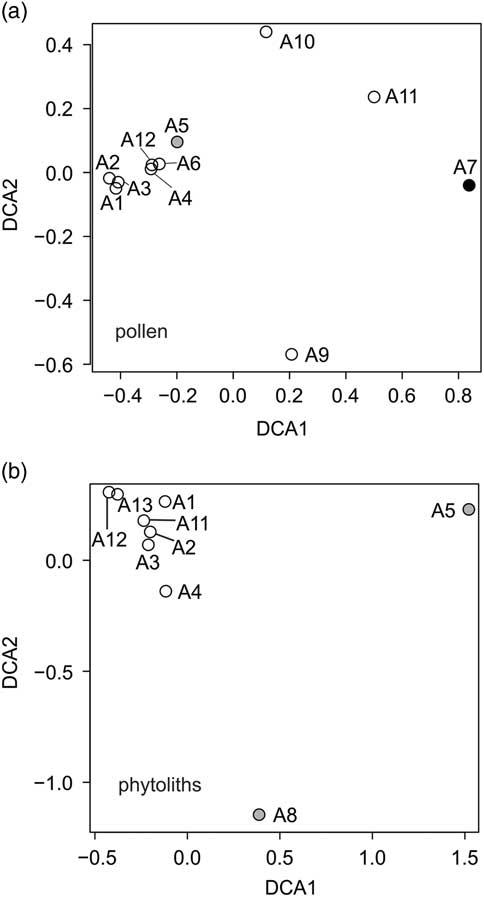
Figure 8 Detrended correspondence analysis of (a) pollen and (b) phytolith data from the Little Blanche Creek arctic ground squirrel midden. Gray circles represent nest material samples containing microtine rodent fecal pellets; other nest material samples are shown as open circles. The black circle represents the sediment sample (A7).
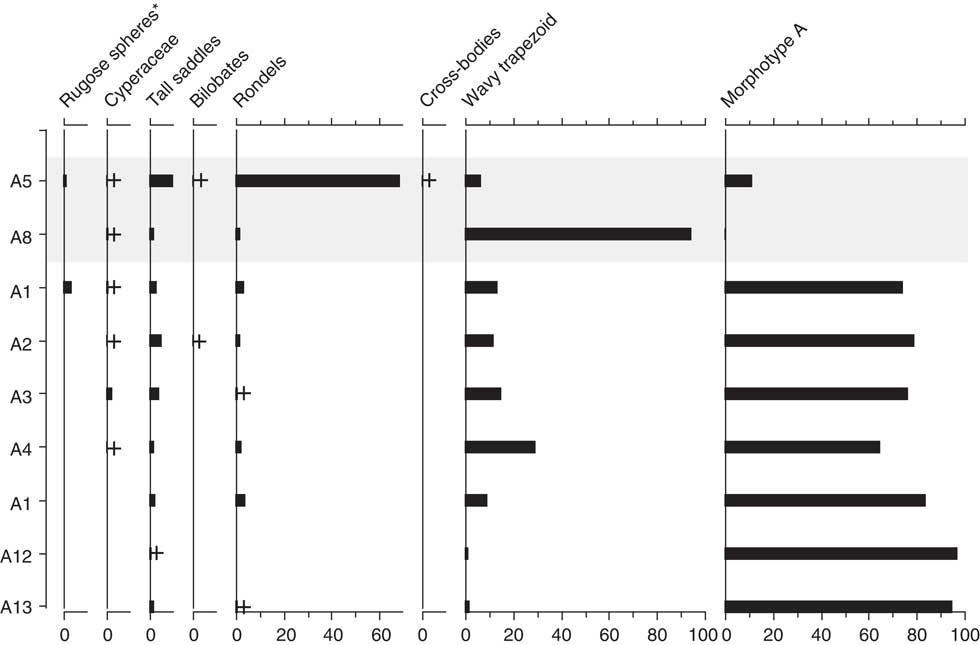
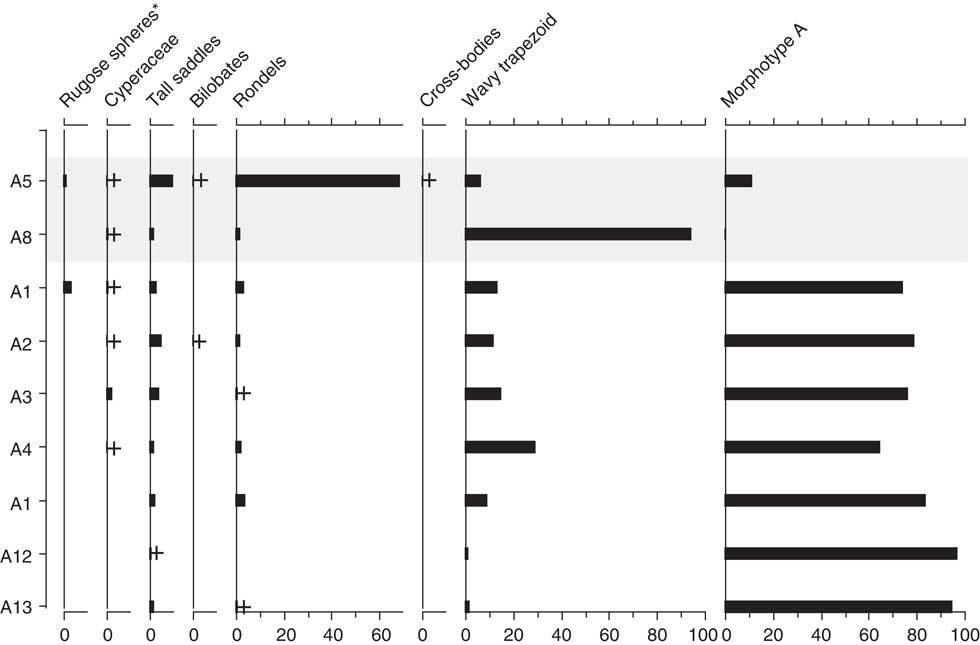
Figure 9 Phytolith assemblages from the Little Blanche Creek midden nest material and nearby sediment. Light gray shading represents nest material samples containing microtine fecal pellets (A5). Samples are listed on the y-axis, and the x-axis is in units of percentage. Plus signs represent presence of a given morphotype in a sample, as in Figure 6. Morphotypes indicating arboreal taxa are marked with an asterisk (*).
Table 3 Taxa preserved as macrofossils in the studied arctic ground squirrel midden.
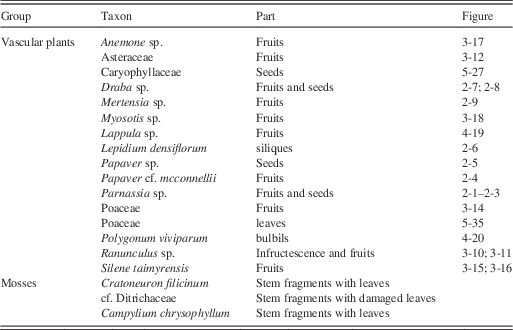
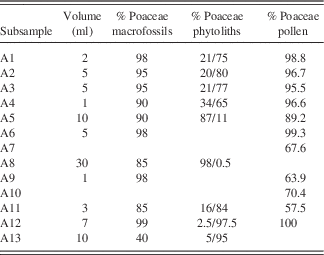
Table 4 Macrofossil sample volume and percentage of poaceous macrofossil material; percentage of poaceous phytoliths; and percentage of poaceous pollen from arctic ground squirrel midden subsamples. The first percentage for phytoliths represents known Poaceae taxa; the second percentage represents an unknown phytolith that may or may not be poaceous.
Microfossils
Pollen
Pollen was generally well-preserved and abundant in the eleven pollen samples. Poaceae dominated the pollen assemblages (Fig. 6), similarly to the macrofossil samples (Table 4). In six samples, some of the Poaceae grains were found still packed together in anther tissue, indicating that they originated from inflorescences that were collected before the release of ripe pollen grains. Other common constituents of the pollen spectra were Artemisia, other Asteraceae, Caryophyllaceae, and Papaver (Fig. 6), albeit in low percentages. Part of the pollen appears more translucent and details such as ornamentation are mostly lacking in subsample A7 (Fig. 7), which consisted of sediment from next to the midden. Except for Poaceae grains, those pollen grains could be easily distinguished from non-reworked pollen grains and were recorded separately as “Reworked pollen.”
The sediment sample (subsample A7) differed from all others in that it contained reworked pollen (Fig. 6). Detrended correspondence analysis (DCA) performed on the pollen assemblages showed that the sediment sample A7 was dissimilar from the nest material samples along DCA Axis 1 (Fig. 8a). Poaceae grains dominated the pollen assemblages for both the sediment and the nest material samples. Artemisia pollen was present in both the sediment and the nest material samples, but was roughly an order of magnitude higher in the sediment sample A7 compared with the nest material samples (Fig. 6). The pollen spectra of A7 contained several forest elements that are either completely absent or present in only one of ten in nest material, such as Alnus, Betula, Picea, and Salix (Fig. 6). Other non-arboreal taxa were present in the sediment sample but similarly absent from nest material samples, such as Asteraceae liguliflorae, Persicaria maculosa type, Polemonium, Saxifraga stellaris type, and the spore of Glomus (Fig. 6). Taxa such as Apiaceae, Caryophyllaceae, Papaver, Plantago, Potentilla type, Ranunculaceae, and a Sporormiella type spore (HdV-113) were present in nest material samples but absent from the sediment sample (Fig. 6). Most of the subsamples of nest material clustered together, though A9, A10, and A11 are compositionally different from the other nest material samples and the sediment sample along DCA Axis 1 and Axis 2 (Fig. 8a). Subsample A5, which was from nest material that contained rodent fecal pellets was not compositionally different from the other nest material subsamples (Fig. 8a).
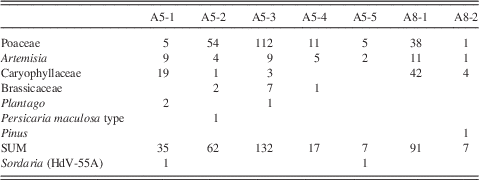
The fecal pellets yielded relatively few pollen grains, and percentage calculations were not possible (Table 5). Poaceae and Caryophyllaceae were the most dominant pollen types, which was similar to the relative pollen percentages in subsample A5 from the nest material (Fig. 6). All of the subsamples of fecal material also yielded Artemisia pollen, which was also present in all but one of the nest material samples and the sediment sample. The other pollen types found in the fecal pellets were also present in at least one of the nest material samples, except a sample from a fecal pellet in A8 which contained Pinus pollen (Table 5).
Table 5 Pollen spectra (numbers) from fecal pellets, probably of microtine rodents, from the arctic ground squirrel midden.
Phytoliths
Poaceae remains were also common in the nine subsamples analyzed for phytoliths (Table 4, Fig. 9). Small percentages of woody taxa (trees) were found in samples A1 (4%) and A5 (2%). Cyperaceae was also found in samples A1, A3, and A8 (Fig. 9). DCA analysis revealed that A5 and A8, which contained the fecal pellets, were very dissimilar to all other samples along Axis 1 (Fig. 8b). The results are dissimilar to the DCA analysis on the pollen signatures (Fig. 8a). An unknown phytolith type, here called Morphotype A (Fig. 5-38), dominated the assemblages (from 65–97.5%) in all samples except A5 and A8, which drove the ordination along DCA Axis 1 (Fig. 8b). When Morphotype A occurred in low percentages or was absent, the assemblage was dominated by either wavy trapezoid, rondel, or saddle morphotypes, indicative of the grass subfamily Pooideae (Fig. 5-39–5-41). The samples where Morphotype A was not dominant were also the samples were fecal pellets were identified and analyzed for pollen. Sample A8 was dominated by wavy trapezoid morphotypes from the Pooideae grasses that drove the ordination of DCA Axis 2 (Fig. 8b).
Non-pollen palynomorphs
Non-pollen palynomorphs were common in all samples, especially a fungal (asco)spore here described as a new type: HdV-834. It is ellipsoidal, one-celled with a smooth thick, light brown wall and simple, slightly protruding apical pores on both ends (Fig. 5-36). Its length averages 36.8 μm (31.5–38.7 μm) and its width averages 18.9 μm (16.9–19.4 μm) (n=10). Its morphology is like a fungal spore illustrated by Pirozynski et al. (Reference Piperno1984: fig. 2c–e), and identified as Arnium sp. Here, we tentatively assign HdV-834 to the order Sordariales. It may represent a dung fungus and be an indicator of herbivores, and the NPP data show that it occurred in high abundances, together with Sordaria-type and Sporormiella-type, but also in the absence of those well-established dung indicators (Davis, Reference Davis1987; van Geel et al., Reference van Geel, Aptroot, Baittinger, Birks, Bull, Cross and Evershed2003; Montoya et al., Reference Miola2010; Gill et al., Reference Giardine, Riemer, Hardison, Burhans, Elnitski, Shah and Zhang2013). Six samples contained Sporormiella and/or Sordaria. The sediment sample A7 is the only sample that contained Glomus chlamydospores (3.3%; Fig. 6).
aDNA
The aDNA yield was high and diverse. Subsample A2 was not visually different from the other subsamples, but was the most productive in terms of intact aDNA sequences. Sample A2 yielded a product with primer pair Spermo CytB 8F+Spermo CytB 132R, which was Sanger sequenced and then BLASTed against and submitted to NCBI GenBank (accession number KY627781). A 100% match with modern Urocitellus parryii CytB sequences from GenBank convincingly confirmed the former presence of an arctic ground squirrel in the midden and validates morphological data (size and contents) pointing to an arctic ground squirrel as the constructor and inhabitant of the midden. Despite its productivity, subsample A2 did not have a higher diversity of identified taxa.
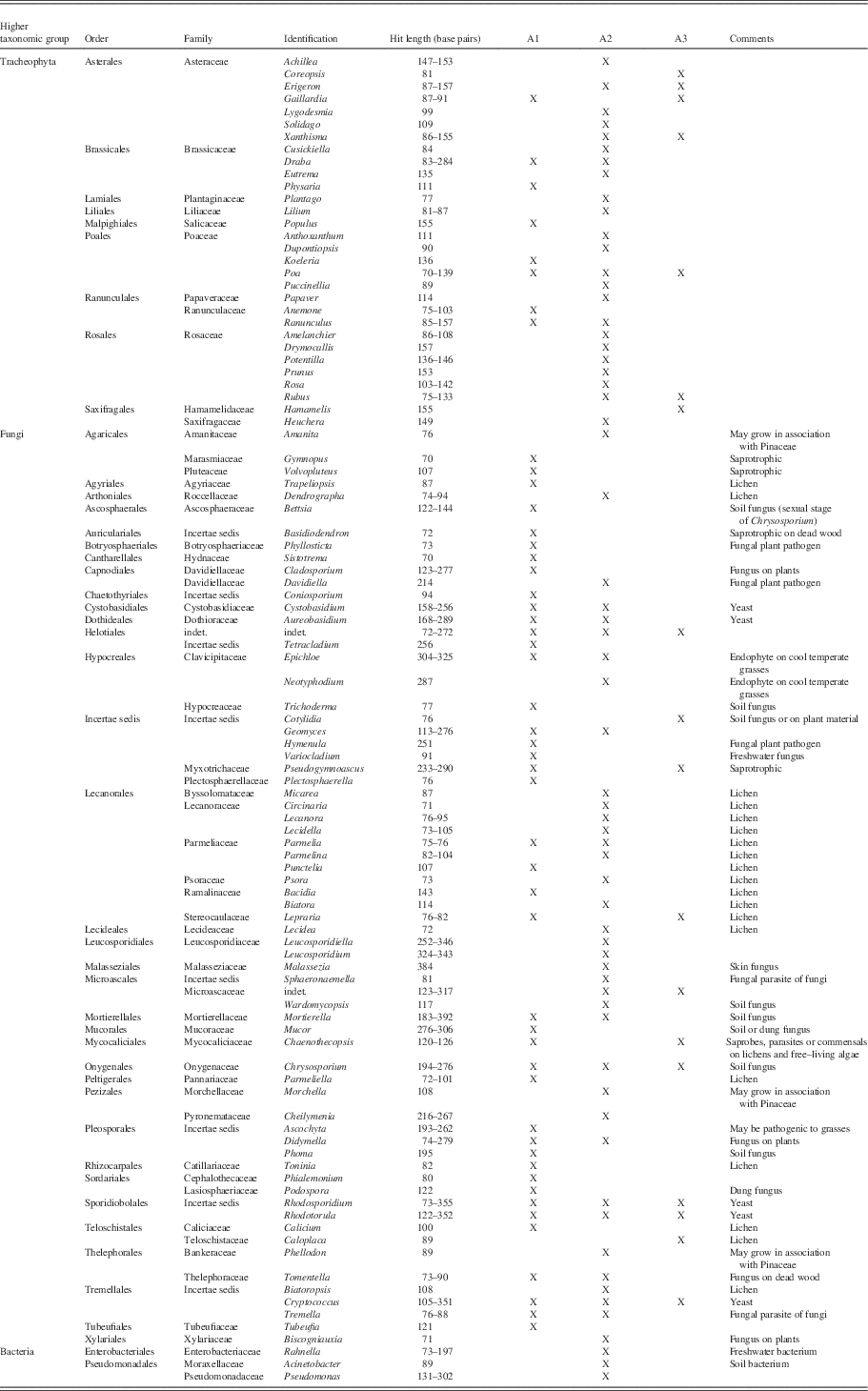
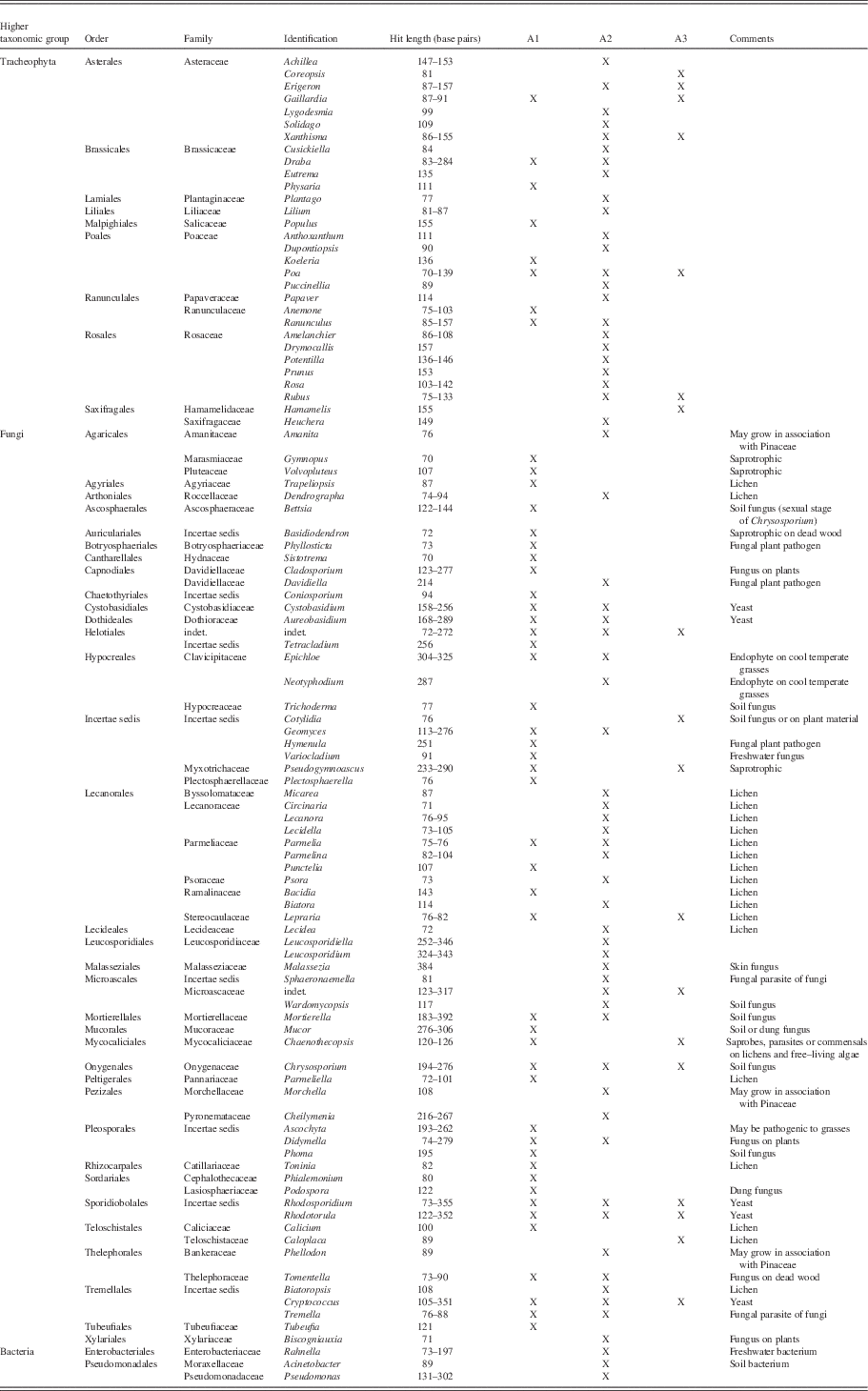
In total, 30 plant genera, 67 fungal genera, and 3 bacterial genera (Table 6) were identified from the aDNA preserved in the midden. Our aDNA data add a significant number of plant taxa identified in ground squirrel middens, including Amelanchier, Coreopsis, Cusickiella, Drymocallis (although considered by some authors to be a synonym of Potentilla), Dupontiopsis, Gaillardia, Hamamelis, Heuchera, Koeleria, Lilium, Lygodesmia, Physaria, Populus, Prunus, Puccinellia, Rosa, Rubus, Solidago, and Xanthisma. Eleven other genera that we recovered through aDNA were already recorded by macrofossil analysis (Table 3; Zazula et al., Reference Zazula, Froese, Elias, Kuzmina and Mathewes2005, Reference Zazula, Froese, Elias, Kuzmina and Mathewes2007). Of the 13 genera identified through macrofossils here, 4 were also retrieved through the aDNA analysis, but 9 were not. Among the fungal aDNA data, we obtained various saprotrophic (including soil) taxa as well as symbionts and vascular plant pathogens, among them parasites on Poaceae in particular, and finally also coprophilous fungi and a skin fungus. We also obtained 19 different lichen taxa.
Table 6 Genera identified from aDNA sequences (trnL/rbcL/nrITS) obtained from the arctic ground squirrel nest. Codes refer to subsamples, “X” marks occurrence.
Discussion
Pleistocene landscapes of Beringia
We used a multi-proxy approach of macrofossils, microfossils, and aDNA from ground squirrel midden in the Yukon region to examine glacial landscapes of Beringia. Our macrofossil analysis from the ground squirrel midden was in broad agreement with Zazula et al. (Reference Zazula, Froese, Elias, Kuzmina and Mathewes2005, Reference Zazula, Froese, Elias, Kuzmina and Mathewes2007), who also studied macrofossil contents of ground squirrel middens from the study area. In addition to taxa previously recorded, we also identified the moss Campylium chrysophyllum in our macrofossil analysis. C. chrysophyllum grows on moist to dry calcareous soils (Lawton, Reference Kołaczek, Zubek, Błaszkowski, Mleczko and Margielewski1971; Crum and Anderson, Reference Crum and Anderson1981) and thus fits with previous reconstructions of a steppe-tundra mosaic as the dominant vegetation during the Late Pleistocene (Zazula et al., Reference Zazula, Froese, Elias, Kuzmina and Mathewes2007). Artemisia pollen was present in almost every sample (Fig. 6), and has been interpreted as a key component of steppe-tundra vegetation. It is also a dominant member of present-day steppe or grassland communities within the subarctic (Edwards and Armbruster, Reference Davis1989). Our fungal analysis showed that lichens were also constituents of the late Pleistocene Beringian ecosystems, supporting previous findings in the region (Lydolph et al., Reference Lawton2005). Our data from the Little Blanche Creek midden add to the scarce literature available on Pleistocene fungi from Eastern Beringia (Pirozynski et al., Reference Piperno1984; van Geel et al., Reference van Geel, Guthrie, Altmann, Broekens, Bull, Gill, Jansen, Nieman and Gravendeel2007), which is a high-latitude fungal biodiversity hotspot (Geml et al., Reference Geml, Laursen, O’Neill, Nusbaum and Taylor2010).
The Little Blanche Creek midden was formed around 30,500 yr ago, a time when most paleoecological reconstructions indicated a steppe-tundra landscape in Beringia (Matthews, Reference Mann, Peteet, Reanier and Kunz1982). Our macro- and microfossil analyses agreed, and grasses were the dominant vegetation of the landscape at the time the midden was formed (Table 4, Fig. 6 and 9). The phytolith data indicated that the Pooideae subfamily grasses were the dominant grass type in the midden material. In the steppe-tundra landscapes, these Pooid grasses are most likely from the genera Calamagrostis, Poa, or Stipa (Blinnikov et al., Reference Blinnikov, Gaglioti, Walker, Wooller and Zazula2011). Macrofossil and aDNA analyses were able to identify 13 and 30 plant genera, respectively, in the local area of the midden (Tables 3 and 6).
Though dominated by grasses and a variety of herbs, all of the microfossil proxies indicated that trees were growing nearby Little Blanche Creek in microrefugia. Phytoliths, which are indicative of local vegetation, showed that these trees were in close proximity to the midden with the presence of tree phytoliths in two of the samples from the nest material (Fig. 9). Our samples contained the fungal genera Phellodon, Amanita, and Morchella, all of which grow in association with Pinaceae (Stalpers, Reference Soreng, Gillespie, Koba, Boudko and Bull1993; Dahlstrom et al., Reference Crum and Anderson2000; Geml et al., Reference Gaglioti, Barnes, Zazula, Beaudoin and Wooller2006). The presence of these fungi in the ground squirrel midden added further evidence for nearby Picea trees in eastern Beringia under full-glacial conditions (Brubaker et al., Reference Brubaker, Anderson, Edwards and Lozhkin2005; Zazula et al., Reference Zazula, Telka, Harington, Schweger and Mathewes2006d). These results were supported by the occurrence of Picea pollen (Fig. 6) in midden material. The aDNA sequence obtained from the ground squirrel nest evidenced that Populus trees also occurred in the nearby microrefugia (compare Mann et al., Reference Lydolph, Jacobsen, Arctander, Gilbert, Gilichinsky, Hansen, Willerslev and Lange2002).
Cusickiella and Dupontiopsis do not occur in Canada in modern times, but were identified through our aDNA analysis. We obtained an aDNA sequence identified as Dupontiopsis at a similarity of 97.78% at a hit length of 90 bp. Soreng et al. (Reference Schoch, Seifert, Huhndorf, Robert, Spougea, Levesque and Chen2015) recently described this grass genus as an alpine endemic to Japan, favoring wet habitats. Soreng et al. (Reference Schoch, Seifert, Huhndorf, Robert, Spougea, Levesque and Chen2015) postulated the ancestor of Dupontiopsis, and the closely allied circumarctic Arctophila and Dupontia, to have lived in Western Beringia. Manual BLASTing of our sequence demonstrated that Dupontia and Arctophila scored just as well as Dupontiopsis. Hence, our sequence may belong to a branch of that clade that once was spread much further eastwards than nowadays and subsequently went extinct.
Representatives of the brassicaceous genus Cusickiella, which we obtained at 98.81 % similarity at a read length of 84 bp, grow on scree and rocky ridges, and are not found further north than the State of Washington (USA) in today’s climate (Flora of North America, Reference Elias2016). Cusickiella may have been able to expand its distribution northward during glacial episodes with more open vegetation and more soil erosion. The identifications of Cusickiella and Dupontiopsis are a further demonstration of the vast biogeographic range changes of plant and animal taxa that occurred in response to climate change during the Late Quaternary and highlight the non-analog character of the flora (Guthrie, Reference Gravendeel, Protopopov, Bull, Duijm, Gill, Nieman and Rudaya1990).
The observed Sporormiella and Sordaria spores and the obtained Podospora aDNA sequence indicate the presence of large herbivores in the region (Fig. 6; van Geel and Aptroot, Reference van Geel2006; Gill et al., Reference Giardine, Riemer, Hardison, Burhans, Elnitski, Shah and Zhang2013). aDNA analyses identified the plant species Eutrema edwardsii in the midden samples, which is strongly nitrophilous (Cody, Reference Cody2000). A high abundance of large herbivores and nitrogen input through deposited dung may have allowed this species to thrive during the Wisconsinan glaciation (Zimov et al., Reference Zimov, Zimov and Chapin2012). Our macrofossil record also contained the fungus Chaetomium, which was recorded by Pirozynski et al. (Reference Piperno1984) from fossil ground squirrel dung from the same area but ca. 10,000 years younger. We also found Malassezia in our samples, which are fungi living on animal skin. The Malassezia that was recorded may have originated directly from the arctic ground squirrel or from other rodents visiting the nest.
Integrating multiproxy paleoecological data
All of the proxies in our analyses provided unique insights into the glacial landscape of Beringia, and together provided a more comprehensive reconstruction than is possible with any single proxy. The use of multiple proxies in our study allowed us to: (1) examine the selective bias of plant material found in ground squirrel middens; (2) compare the local versus regional input of macro- and microfossils into ground squirrel middens; (3) assess factors such as under- and over-representation and variability of taxa; and (4) improve the taxonomic resolution of remains found in the ground squirrel midden. The integration of datasets such as the ones used here will assuredly improve reconstructions of past landscapes from midden material, and also from other paleoecological archives.
Even though tree taxa were present in pollen assemblages, they are all genera with wind-dispersed pollen grains, meaning they could have originated from trees far away from the midden (i.e., long distance transport). Phytoliths indicate local vegetation; they are deposited into soils at the site of plant decay. Thus, the phytoliths found in the midden (Fig. 9) were from locally derived plants. The presence of tree phytoliths indicates that a microrefugium harboring trees was present near the Little Blanche Creek midden during the Wisconsinan glaciation. The complementarity of the pollen, spores, and aDNA documented that these tree assemblages were composed of Populus and Picea.
Abundances of macro- and microfossils from midden materials are difficult to interpret in terms of vegetation composition because of the selection made by ground squirrels (Zazula et al., Reference Zazula, Mathewes and Harestad2006a; Fig. 6 and 9) and the variability in productivity of fossil material between taxa. For example, some plants are known to produce more pollen than others in relation to their basal area within the landscape (Davis, Reference Dahlstrom, Smith and Weber1963; Jacobson and Bradshaw, Reference Hofreiter, Poinar, Spaulding, Bauer, Martin, Possnert and Pääbo1981). Poaceae are known to produce large amounts of pollen. The occurrence of some Poaceae grains still packed together in anther tissue demonstrated that Poaceae pollen in our samples is over-represented, which has also been shown in other studies from this setting (Gravendeel et al., Reference Goecks, Nekrutenko and Taylor2014). Because Poaceae are also prolific phytolith producers, we are unable to verify the over-representation of Poaceae pollen in our samples.
High abundances of reworked pollen and Glomus chlamydospores (Fig. 6), which are formed below the soil surface (Anderson et al., Reference Anderson, Homola, Davis and Jacobson1984; Kołaczek et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock and Buxton2013), were found in the A7 sediment sample. The reworked pollen indicates that the sediment sample likely encompasses a time span larger than the microfossils found in nest material samples. Reworked Sphagnum spores were also found in A7. These may be significantly older than any of the other pollen and spores, as they do not fit, from an ecological viewpoint, in the obtained microfossil spectra. As a whole, these results demonstrate that sediments or loess deposits nearby middens should be analyzed in conjunction with midden material to provide a more comprehensive view of past vegetation patterns, keeping in mind the age difference between the midden itself and (the older) surrounding sediment due to the animal digging down.
The macro- and microfossil analyses were in broad agreement regarding the taxa present in the sediment sample, nest material, and fecal pellet samples (Table 4, Fig. 6 and 9). The DCA analysis of pollen and phytoliths, however, showed separations of samples differently (Fig. 8). For example, the A5 sample had a similar composition to other nest material samples in the pollen DCA, but was very dissimilar to other nest material samples in the phytolith DCA (Fig. 8). This difference actually shows the complementarity of the proxies to provide a more robust reconstruction than either proxy alone. The pollen assemblages can more reliably distinguish tree and herbaceous taxa, whereas the phytoliths are more sensitive to detecting changes in the composition of grass communities (Piperno, Reference Morcote-Ríos, Giraldo-Cañas and Raz2006; Morcote-Ríos et al., Reference Montoya, Rull and van Geel2015). A combined DCA that incorporated both proxies was not possible in our analysis because there were too few overlapping samples. Also, the variation of the pollen spectra from the fecal pellets suggested slight differences in the diet of the microtine rodents. These pellets may not have been dropped simultaneously, and suggest repeated visits by microtine rodents to the arctic ground squirrel midden.
The high yield of aDNA shows the significant potential of the Pleistocene arctic ground squirrel middens as sources of species or genus-specific paleoecological reconstructions. Of the 19 previously unrecorded genera identified via aDNA analyses, 9 currently occur in the Yukon Territory (Cody, Reference Cody2000; VASCAN, Reference van Geel, Zazula and Schweger2016). Of the 10 that do not, 8 are currently found elsewhere in Canada (VASCAN, Reference van Geel, Zazula and Schweger2016). Most of the newly recorded genera are members of the large and complex families Asteraceae (5 genera), Poaceae (3), and Rosaceae (5), which are many times indistinguishable using traditional macrofossil or microfossil analyses. These results highlight the advantage of including aDNA as a proxy in paleoecological reconstructions when possible. The NCBI GenBank and the UNITE Fungal ITS Database are continuously growing and therefore future analyses with aDNA from paleoecological archives will increase in potential to identify macro- and microfossil remains at the genus or species level. aDNA from an array of plants and animals (e.g., Elias, Reference Elias and Brigham-Grette2010) in paleoecological archives such as middens are proving valuable in addressing a range of evolutionary and ecological questions. Additional work, specifically with plant aDNA, will undoubtedly reveal additional information about past plant migrations, further detailing the non-analog nature of glacial environments in this region and strengthening the notion of Beringia as a glacial biotic refuge in the Arctic.
Conclusions
Here, we used a multiproxy analysis of macrofossils, pollen, fungal spores, phytoliths, and aDNA from a ground squirrel midden to provide information on the glacial landscapes of Beringia. The data generated from the multiproxy approach provided complementary information that provided a more robust reconstruction than any of the individual proxies alone. We also showed that combining the proxies allowed a more in-depth interpretation of regional versus local signals of microfossil assemblages, but also that several subsamples are needed for reliable results. We identified a total of 39 vascular plant and bryophyte and 68 fungal genera from the arctic ground squirrel midden macrofossil and aDNA records. Future studies incorporating a multi-proxy approach of macrofossils, microfossils, and aDNA will add a wealth of additional information on the glacial vegetation in Eastern Beringia. But this approach will also benefit paleoecological studies in other time periods and other geographic regions.
Acknowledgments
Placer gold miner Miles Carlson allowed us to sample the artic ground squirrel midden and provided guidance and hospitality out in the Klondike goldfields. Dr. Raymond van der Ham (Naturalis Biodiversity Center, The Netherlands) contributed to discussions. Annemarie Philip (Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, The Netherlands) prepared the pollen slides. Jan van Arkel (IBED) prepared photographs and plates; Elza Duijm (NBC) assisted with work on aDNA. Dr. Jacqueline van Leeuwen (University of Bern, Switzerland) helped with identification of some pollen types. Dr. Alberto Reyes suggested literature. Elizabeth Hall and Susan Hewitson (both Yukon Palaeontology Program, Canada) assisted our work at the field site. Thanks to all.
Supplementary material
For supplementary materials referred to in this article, please visit https://doi.org/10.1017/qua.2017.93