Introduction
Trypanosomes are protozoan parasites that are distributed globally which infect humans, vertebrate animals and intermediate invertebrate hosts. Among them, members of the subgenus Herpetosoma, such as Trypanosoma lewisi and Trypanosoma musculi, are commonly found in rodents (Hoare, Reference Hoare1972; Kostygov et al., Reference Kostygov, Karnkowska, Votýpka, Tashyreva, Maciszewski, Yurchenko and Lukeš2021). These 2 trypanosomes cannot be easily distinguished due to their high degree of similarity in morphological characteristics and genetic markers such as the SSU rDNA sequences (Hong et al., Reference Hong, Zhang, Fusco, Lan, Lun and Lai2017). However, they do significantly differ in many aspects. In particular, T. lewisi infects only rats and sometimes humans (Sarataphan et al., Reference Sarataphan, Vongpakorn, Nuansrichay, Autarkool, Keowkarnkah, Rodtian, Stich and Jittapalapong2007; Verma et al., Reference Verma, Manchanda, Kumar, Sharma, Goel, Banerjee, Garg, Singh, Balharbi, Lejon, Deborggraeve, Singh Rana and Puliyel2011), while T. musculi infects only mice and is unlikely to be pathogenic to humans (Zhang et al., Reference Zhang, Hong, Li, Lai, Hide, Lun and Wen2018). A few research studies have indicated that both T. musculi and T. lewisi can modulate the host immune response in coinfections with various other infectious agents, to potentially cause more harm to the hosts, by altering the infection kinetics and increasing the duration of colonization in the host (Lowry et al., Reference Lowry, Leonhardt, Yao, Belden and Andrews2014; Vaux et al., Reference Vaux, Schnoeller, Berkachy, Roberts, Hagen, Gounaris and Selkirk2016; Nzoumbou-Boko et al., Reference Nzoumbou-Boko, De Muylder, Semballa, Lecordier, Dauchy, Gobert, Holzmuller, Lemesre, Bras-Gonçalves, Barnabé, Courtois, Daulouède, Beschin, Pays and Vincendeau2017; Gao et al., Reference Gao, Yi, Geng, Xu, Hide, Lun and Lai2021). To gain a better understanding of the biological characteristics, the kinetoplast DNA (kDNA) of T. lewisi has been comprehensively analysed (Lin et al., Reference Lin, Lai, Zheng, Wu, Lukeš, Hide and Lun2015; Li et al., Reference Li, Zhang, Lukeš, Li, Wang, Qu, Hide, Lai and Lun2020). However, little is known of the details of the kDNA in T. musculi.
Trypanosomes are members of the kinetoplastea group of protozoa, named due to the presence of the kDNA. Trypanosome kDNA is a specific network structure of interlocking mitochondrial DNA circles, which consists of thousands of minicircles with dozens of maxicircles (Lukes et al., Reference Lukes, Guilbride, Votýpka, Zíková, Benne and Englund2002). Earlier research on Trypanosoma brucei has shown that kDNA comprises at least 5% of the total cellular DNA, while most other eukaryotic mitochondrial DNA accounts for no more than 1% (Lukeš et al., Reference Lukeš, Wheeler, Jirsová, David and Archibald2018). In general, kDNA maxicircles encode functional homologues of mitochondrial genes which are flanked by non-coding regions that diverge significantly in sequence and size amongst trypanosome species (Simpson et al., Reference Simpson, Neckelmann, de la Cruz, Simpson, Feagin, Jasmer and Stuart1987; Sloof et al., Reference Sloof, de Haan, Eier, van Iersel, Boel, van Steeg and Benne1992; Westenberger et al., Reference Westenberger, Cerqueira, El-Sayed, Zingales, Campbell and Sturm2006). One of the unusual features of the kinetoplast is that most of the maxicircle gene transcripts are not mature and do not encode functional open reading frames. These encrypted transcripts become translatable only after post-transcriptional processing, namely RNA editing, that inserts and deletes uridine residues (Stuart et al., Reference Stuart, Allen, Heidmann and Seiwert1997, Reference Stuart, Schnaufer, Ernst and Panigrahi2005). RNA editing was first discovered in cytochrome oxidase subunit 2 (COII) gene of Trypanosoma brucei and Crithidia fasciculata, whose mRNA transcripts have 4 uridine insertions (Benne et al., Reference Benne, Van den Burg, Brakenhoff, Sloof, Van Boom and Tromp1986). The minicircles, recognized by a conserved motif of 12 nucleotides (GGGGTTGGTGTA) (Ray, Reference Ray1989), encode guide RNA (gRNA) molecules that accurately position the editing machinery to ensure correct maxicircle transcripts are produced (Blum and Simpson, Reference Blum and Simpson1990).
Here, using PacBio and Illumina sequencing reads, the complete maxicircle sequence of T. musculi was assembled and annotated, including the repetitive non-coding variable region. Comparative analyses indicate that the gene organization and distribution in T. musculi maxicircles are highly conserved with T. brucei, T. cruzi and T. lewisi. The maxicircle kDNA gene organization of T. musculi and comparison with its species relatives was also presented. In addition, the genetic information on the divergent region (DR) II reveals that it may provide a good marker for molecular diagnosis and molecular epidemiological investigation of trypanosomes.
Materials and methods
Parasites, ultrastructure, kDNA extraction and restriction endonuclease digestion
Trypanosoma musculi Partinico II strain was gifted by Professor Philippe Vincendeau of Université de Bordeaux, France, which was originally obtained from the London School of Hygiene and Tropical Medicine (Krampitz, Reference Krampitz1969). T. musculi Particino 2, Lincicome and CDC strains were purchased from American Type Culture Collection (ATCC). Trypanosomes were harvested from the blood of infected mice and cultured in RPMI-1640 medium at 37°C supplemented with 10% fetal bovine serum (FBS) and a feeder layer of mouse macrophages as modified from Behr (Behr et al., Reference Behr, Mathews and D'Alesandro1990). Protocols for the use of mice were approved by the Institutional Review Board for Animal Care at Sun Yat-Sen University under license 31672276. For transmission electron microscopy, trypanosome specimens were prepared according to the method of Bozzola (Bozzola, Reference Bozzola and Kuo2014), and observed by using the JEM-100CX-II microscope system. For T. musculi DNA preparations, total DNA was purified using a phenol–chloroform method and kDNA was extracted by sucrose gradient ultracentrifugation according to previously published methods (Pérez-Morga and Englund, Reference Pérez-Morga and Englund1993). The isolated kDNA network was visualized on a 1% agarose gel and analysed with restriction enzymes HindIII, EcoRI, BamHI, RsaI, HaeIII and TaqI (New England Biolabs, USA). A computer-simulated restriction enzyme digestion map of T. musculi maxicircle was generated using the Dnaman 9.0 software (Lynnon Corporation, Quebec, Canada) based on the sequence assembled in this study.
Immunofluorescence assay
Trypanosome cells (1 × 107 cells mL−1) were centrifuged for 5 min at 3000 × g and washed twice in phosphate-buffered saline (PBS). The cells were then transferred onto clean slides, which were left to air-dry in a fume hood, following fixation by methanol for 10 min. Dried slides were rehydrated and washed twice in PBS for 5 min at room temperature. The slides were then incubated with primary mAb-anti-L8C4 (1:800) followed by incubation with Cy3-Conjugated goat anti-mouse IgG (A10521, Thermo Fisher) (1:400) followed by counterstaining consisting of 1 × PBS with 3 μg mL−1 4,6-diamidino-2-phenylindole (DAPI) (Kohl et al., Reference Kohl, Sherwin and Gull1999). They were then photographed using a Leica fluorescence microscope.
Deep sequencing, sequence assembly and PCR verification
To generate a high-quality maxicircle assembly, a kDNA Illumina library was constructed and sequenced using Illumina HiSeq2000 technology commercially (Novogene, China). Also, a PacBio Sequel library was constructed using total DNA and sequenced commercially (Annoroad, China). The Illumina reads were checked for quality and trimmed using fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic (Bolger et al., Reference Bolger, Lohse and Usadel2014). Canu 2.0 software was used in de novo assembly of the T. musculi genome with parameter ‘genomeSize = 30 m minReadLength = 600 minOverlapLength = 300 corOutCoverage = 100 corMinCoverage = 2 correctedErrorRate = 0.035’ using the PacBio reads (Koren et al., Reference Koren, Walenz, Berlin, Miller, Bergman and Phillippy2017). Then, the genome contigs, assembled from PacBio reads, were polished using the Illumina reads by Pilon software to improve genome assembly (Walker et al., Reference Walker, Abeel, Shea, Priest, Abouelliel, Sakthikumar, Cuomo, Zeng, Wortman, Young and Earl2014). Finally, the assembly sequences were aligned, with BLAST, to a previously obtained T. lewisi maxicircle assembly (KR072974) and redundant overlap deleted by MEGA 7.0 to yield the complete maxicircle sequence (Camacho et al., Reference Camacho, Coulouris, Avagyan, Ma, Papadopoulos, Bealer and Madden2009; Kumar et al., Reference Kumar, Stecher and Tamura2016). To obtain T. b. rhodesiense, Trypanosoma grayi and T. lewisi complete maxicircle genomes, processed reads were assembled from WGS data (SRX3199071, SRX620256 and SRR11918574, respectively) freely available on NCBI using SPAdes 3.12.0 with parameter ‘--plasmid --careful -t 16 -m 200’ (Antipov et al., Reference Antipov, Hartwick, Shen, Raiko, Lapidus and Pevzner2016). Then, alignment and trimming were also carried out as completed for T. musculi. The sequences of the T. musculi maxicircle coding region were also corrected by PCR verification using 12 pairs of primers. Meanwhile, MURF2, ND4 and ND5 genes were also amplified in 3 additional T. musculi strains (T. musculi Particino 2, Lincicome and CDC). Primers used are summarized in Table S1.
Gene annotation
Annotation of T. musculi maxicircle coding regions was performed by comparison with T. brucei (EATRO 427, M94286.1), T. cruzi (CL, DQ343645.1) and T. lewisi (CPO02, KR072974.1) manually using BLAST. Patterns of RNA editing of T. musculi maxicircle genes were predicted according to GC% and RNA editing pattern of T. lewisi (Li et al., Reference Li, Zhang, Lukeš, Li, Wang, Qu, Hide, Lai and Lun2020).
Data analysis
Dotplot graphs of the T. musculi maxicircle sequence plotted against itself and 3 other Trypanosomatidae species were generated by YASS software with default parameter (allow 10% indels, 25% mutations and e-value <1 × 10−5 in alignment) (Noé and Kucherov, Reference Noé and Kucherov2005). GC percentage, assembly coverage and homology search algorithms were drawn using Circos v0.69 (Krzywinski et al., Reference Krzywinski, Schein, Birol, Connors, Gascoyne, Horsman, Jones and Marra2009). Regions (>300 bp) with sufficient sequence identity were plotted as coloured ribbons, denoting the percentage of sequence identity. BioEdit software was used to create alignments and calculate nucleotide percentage identity matrices among different Trypanosomatidae species (Hall, Reference Hall1999). Curation of the palindromes and inserted sequence homology analysis was performed using BLAST. MEME software was used to identify motifs and generate LOGO diagrams in the DR I region (Bailey et al., Reference Bailey, Johnson, Grant and Noble2015).
Phylogenetic analysis
The entire coding region of the kinetoplast maxicircles were aligned using ClustalO 1.2.4 (Sievers et al., Reference Sievers, Wilm, Dineen, Gibson, Karplus, Li, Lopez, McWilliam, Remmert, Söding, Thompson and Higgins2011) and the alignment was trimmed using Gblocks 0.91b with option ‘−t = d -b4 = 5 -b5 = h’ (Talavera and Castresana, Reference Talavera and Castresana2007). Maximum likelihood trees were generated by RAxML 8.2.12 with 1000 bootstrap replicates (Stamatakis, Reference Stamatakis2014). Neighbour joining and Minimum evolution trees were performed using MEGA 7.0 including 1000 bootstrap pseudo-replicates. Maxicircle genome sequences used in this work are summarized in Table S2.
Results
Morphology, ultrastructure, kDNA isolation and restriction enzyme digestion
In culture, T. musculi cells tend to attach to each other and form a rosette-like pattern (Fig. 1A), via their flagella, as determined using specific antibodies against the paraflagellar rod (Fig. 1C). At this stage, T. musculi is at the epimastigote stage in which the kinetoplast lies closely beside the nucleus. Ultrastructural analysis showed that the kinetoplast DNA disc was 660 ± 99 nm in length and 152 ± 26 nm in width (n = 50) (Fig. 1B), which is similar to closely related species such as T. lewisi (Lin et al., Reference Lin, Lai, Zheng, Wu, Lukeš, Hide and Lun2015).

Fig. 1. Morphology and ultrastructure of T. musculi. (A) Giemsa staining of the epimastigote form of T. musculi from in vitro culture (scale bar 5 μm). Flagellum (F), nucleus (N) and kinetoplast (K) are indicated. (B) Electron micrograph of the T. musculi trypomastigote form (scale bar 200 nm). Basal body (BB), flagellum (F), flagellar pocket (FP) and kinetoplast (K) are indicated. (C) Immunofluorescence analysis (IFA) shows epimastigote-like forms of T. musculi from in vitro cultivation with antibody L8C4 detecting paraflagellar rods (green) and DAPI detecting kinetoplast DNA and nuclear DNA (blue) (scale bars 5 μm).
A total of 1010 T. musculi were harvested and high-quality kDNA was obtained with a 260/280 absorbance ratio of 1.80. Kinetoplast DNA was found to be intact and free from contamination with nuclear or host DNAs as judged by agarose electrophoresis (Fig. S1A). Meanwhile, kDNA was incubated with endonucleases of HindIII, EcoRI, BamHI, RsaI, HaeIII and TaqI and a computer-simulated restriction enzyme digestion map was generated based on the maxicircle assembly which is described later (Figs S1B–C). Some bands smaller than 4.0 kb did not correspond to the computed simulated patterns of T. musculi maxicircle, which implies the presence of a high number of possible heterogeneous minicircles in the kDNA of T. musculi. The bands consistently observed in Fig. S1B at ~1.3 kb suggest the presence of minicircles of a similar size as reported in T. lewisi (Li et al., Reference Li, Zhang, Lukeš, Li, Wang, Qu, Hide, Lai and Lun2020). Patterns that are free of kDNA in the wells were achieved using HaeIII and TaqI and indicated a high frequency of cleavage of kDNA minicircles. Most likely, the bands with molecular sizes of >4 kb correlated with the computer-simulated patterns (Fig. S1C), are derived from kDNA maxicircles, except a band (~5 kb) in RsaI lane, a potential result of incomplete digestion. Moreover, the presence of 4 large molecular weight bands, in the EcoRI digestion, with sizes of >10, ~7, ~6 and ~4 kb, indicated that the full-size kDNA maxicircle is larger than the sum of 27 kb.
Assembly and annotation of the kDNA maxicircle
Genomic DNA from the T. musculi Partinico II strain was sequenced on PacBio Sequel and Illumina platforms and contigs were assembled with the long-read assembler Canu 2.0 and corrected with the Illumina reads in Pilon. Then, a contig in length of 38 603 bp was identified in a BLAST search against the T. lewisi maxicircle (KR072974). This had 2 overlapping regions of 4002 bp (covering positions from −3341 or 31 266 to 661 bp) at each end (Fig. S2) confirming completion of the circle. Meanwhile, the maxicircle sequence has also been confirmed using 5 overlapping raw reads from the PacBio library (Fig. S2) and the maxicircle coding region sequences were also further refined using 12 pairs of primers and Sanger sequencing validation (Fig. S3). Finally, a 34 606 bp-long complete T. musculi maxicircle sequence was obtained, with an average coverage of 13.2X from Illumina reads and 268X from PacBio reads, including the coding regions (16 975 bp) and the DRs (17 631 bp). The overall GC content of the maxicircle was 23.7%, with 27.5% in coding regions and 20.1% in DRs (Fig. 2).
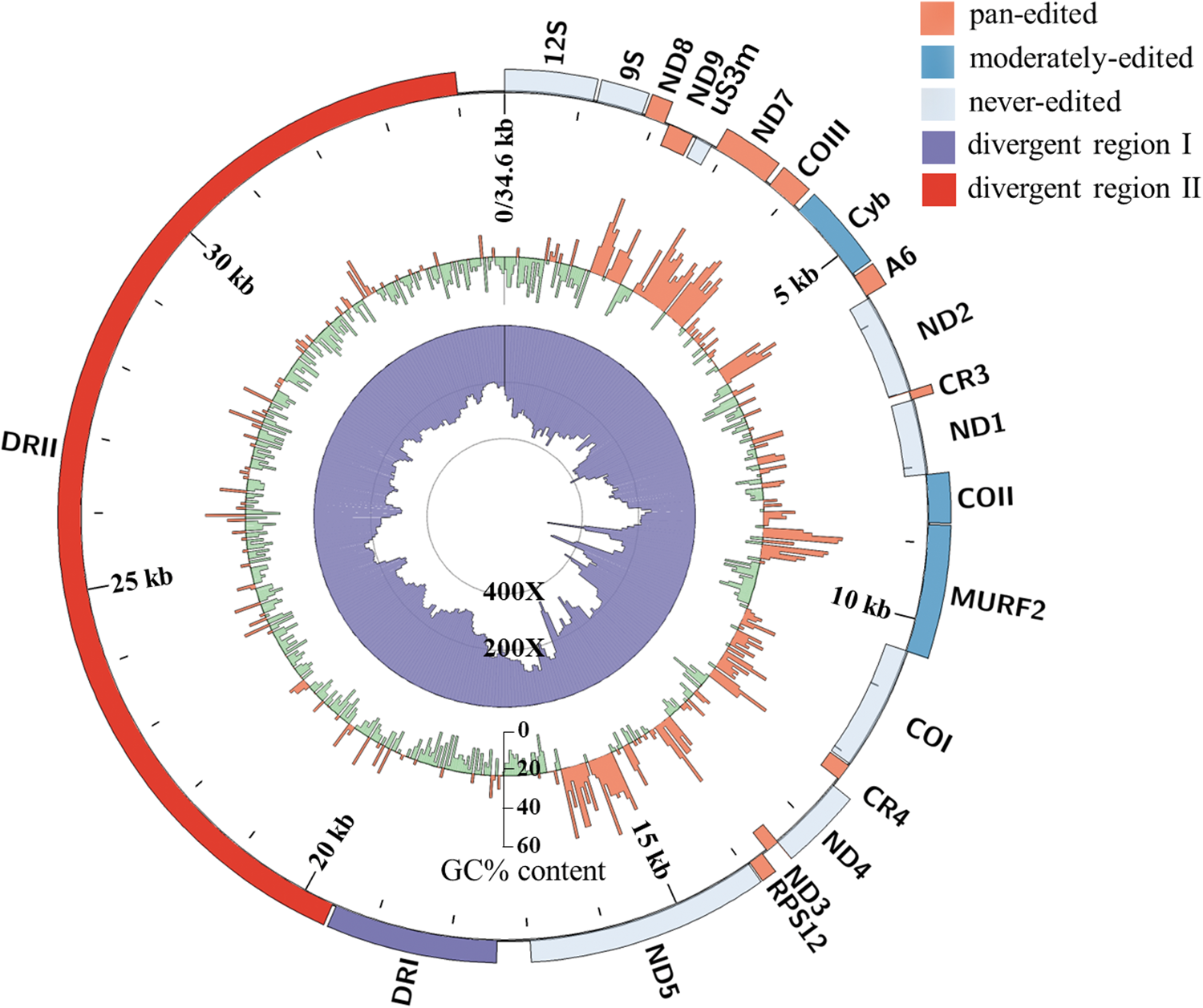
Fig. 2. Circos plot of the T. musculi maxicircle. The outer track indicates the gene arrangements and gene distribution; the middle track represents the GC content (orange for above-average and green for below-average) and the inner track is a histogram of assembly coverage.
Twenty genes were annotated in the T. musculi maxicircle by comparison with known Trypanosomatidae species (T. brucei, T. cruzi and T. lewisi), as listed in Table 1. All genes were found to be syntenic with the maxicircles of the comparator Trypanosomatidae species T. brucei, T. cruzi and T. lewisi (Fig. 2, Fig. S4). A sequence homology analysis (Fig. S5), showed that the T. musculi maxicircle has 92.7% identity to T. lewisi (blue ribbons). The ribbons change to yellow when compared with T. cruzi (78.0% identity) and T. brucei (73.9% identity), largely due to the low similarity in extensively edited genes (Table 2). Moreover, 3 breaks shown as discontinuations of lines or ribbons (Figs S4 and S5) appear in T. musculi genes MURF2, ND5 and ND4, with 2 sections (630 and 1278 bp) inserted and 1 section deleted (281 bp), respectively (Fig. 3A and Table 1).

Fig. 3. Analysis of insertions or deletions in the T. musculi maxicircle genes MURF2, ND5 and ND4. (A) A diagram of insertions or deletions in the T. musculi maxicircle genes MURF2 (630 bp insertion), ND5 (1278 bp insertion) and ND4 (281 bp deletion). Insertion regions are labelled as grey boxes. (B) PCR amplification of the T. musculi MURF2, ND5 and ND4 genes from T. musculi Partinico II (1), T. musculi Particino 2 (2), T. musculi Lincicome (3), T. musculi CDC (4), and fragments are analysed on a 1.0% agarose gel. M, DL2000 marker (Takara, China). N, negative control. The positions of primers are shown in (A) and Table S1. (C) Alignment of the conserved regions from the T. lewisi minicircle (MN447335.1) and insertion sequences of MURF2 and ND5. Conserved sequence blocks (CSB-I, II, III) are highlighted in grey.
Table 1. Gene positions and lengths in the maxicircle of T. musculi

Gene positions are shown relative to the start of the 12S rRNA.
a These genes are encoded on the reverse strand.
b CR3 2 end positions from T. musculi, T. lewisi, T. cruzi and T. brucei are uncertain.
c A fragment deletion is found in the T. musculi ND4 gene.
Table 2. Comparison of the average percentage identities of T. musculi kDNA maxicircle with those from the other 3 Trypanosomatidae species

Entire coding region: starting from 5′end of 12S rRNA to 3′ end of ND5.
5′-edited genes: Cyb, COII.
Extensively edited genes: ND8, ND9, ND7, COIII, ATPase6, CR3, CR4, ND3, RPS12.
Non-edited genes: uS3m, ND2, ND1, COI.
MURF2, ND4 and ND5 genes are not calculated in T. musculi (5′-edited genes or non-edited genes) due to insertions/deletion.
The confirmation of mutations in MURF2, ND5 and ND4 was performed with 3 other strains (T. musculi Particino 2, Lincicome and CDC). PCR results showed that the insertion in MURF2 is specific to T. musculi Partinico II strain and not present in other 3 strains, while the insertion in ND5 and the deletion in ND4 exist in all tested strains (Fig. 3B). Furthermore, these insertions and the deletion have also been confirmed by inspecting alignments of the raw reads mapped back to the maxicircle assembly. Alignment analysis of insertions showed that a fragment (150 bp) at the 5′ end region of MURF2 insertion sequence is homologous to both the 5′ end and middle regions of ND5 insertion sequence (Fig. 3C). Moreover, those sequences in MURF2 and ND5, respectively, share 95.3, 96 and 95.3% identity with conserved regions of T. lewisi minicircles (MN447336.1, MN447339.1 and MN447386.1), and these 150 bp homologous regions cover 3 conserved sequence blocks (CSBs) of minicircles, indicating a minicircle origin of both insertions in MURF2 and ND5. Together with the data shown in Fig. S1 and Fig. 3C, it seems that, unsurprisingly, T. musculi has a similar size and structure to minicircles reported for T. lewisi (Li et al., Reference Li, Zhang, Lukeš, Li, Wang, Qu, Hide, Lai and Lun2020), i.e. ~1.3-kb category I minicircles that have 2 conserved regions with CSB1-3 motifs (and perhaps also ~1.5 kb category II minicircles with only 1 conserved region; such a band is also apparent in Fig. S1B). It therefore appears that the ~1.3 kb ND5 insertion corresponds to a (degenerated) category I minicircle and the 630 bp MURF2 insertion corresponds to half a category I minicircle.
RNA editing patterns of the maxicircle have been well studied in T. brucei and T. lewisi (Gerasimov et al., Reference Gerasimov, Gasparyan, Kaurov, Tichý, Logacheva, Kolesnikov, Lukeš, Yurchenko, Zimmer and Flegontov2018; Li et al., Reference Li, Zhang, Lukeš, Li, Wang, Qu, Hide, Lai and Lun2020) and they are well correlated with GC%. The GC% pattern in T. musculi is fairly similar to T. brucei and T. lewisi. However, unexpectedly high GC contents were noticed in MURF2 and ND5 (Fig. S6), and they are precisely attributed to the insertions in both genes. In another region, COII and its cis-acting gRNA were identified (Table 1).
The whole coding region of the maxicircle is considered as a valuable marker for phylogenetic relationships of Trypanosomatidae species (Kaufer et al., Reference Kaufer, Stark and Ellis2019). To further confirm the evolutionary relationship of T. musculi and other Trypanosomatidae species, sequences corresponding to whole coding region of T. musculi were aligned with the sequences from other Trypanosomatidae species to infer phylogenetic relationships. In the tree, T. musculi and T. lewisi are identified as belonging to the same subgenus Herpetosoma, clustered with the sister groups of Schizotrypanum and Aneza (Fig. 4).

Fig. 4. Phylogenetic analysis of the Trypanosomatidae species using the maxicircle coding regions. Phylogenetic tree is performed based on Maximum likelihood /Neighbour joining/Minimum evolution methods with 1000 bootstrap replicates with the respective bootstrap confidences indicated at branch points. Branch lengths are indicated by the black line and the scale bar represents the number of nucleotide substitutions per site.
Sequence analysis of the maxicircle DRs
A common theme of the maxicircle DR is the presence of various repeat arrays, which is also the case for T. musculi. The full map of DR of T. musculi was built by the YASS and Circos packages to identify homologous regions and to show global patterns of DR organization (Fig. 5A and B). Dot-plot analyses of the DR showed 2 typical sections (I, II), flanking either the 12S rRNA or ND5. DR I is in a length of about 1.6 kb, which is composed of short and highly repetitive units of about 107 bp, with 2 motifs being found (Fig. 5C). While DRII is in a length of about 14 kb, it consists of a series of tandem elements, namely α, β, γ, σ, and short version α′, β′, γ′ (Fig. S7).

Fig. 5. Sequence analyses of the DR of the T. musculi maxicircle. (A) Dot-plot analysis of the T. musculi maxicircle, the main diagonal line represents the sequence's alignment with itself and the lines about the main diagonal represent repetitive patterns within the maxicircle sequence. (B) Circos plot of the DR of T. musculi maxicircle, the outer track indicates gene arrangement and gene distribution. Ribbons inside the circle connect homologous regions, colour represents per cent of sequence identity in the range (70%; 100%) in the order yellow, green and blue. Four palindromes are shown with red arrows. (C) The repetitive sequences from DR I are aligned and 2 motifs predicted by MEME are indicated with a black line. LOGO diagrams show nucleotides at a given position of each motif and relative frequency represented by height. (D) The palindromes from the DR of the Trypanosomatidae species are shown with the palindromic bases highlighted.
Palindromes are a typical structure already found in T. cruzi, T. lewisi and T. rangeli. Based on identifying homologues using BLAST, 4 AT-rich conserved palindromes showed up in the DRII (Fig. 5B and D). Palindromes I and IV have the same perfect palindrome structure, 34 bp long, and are located at 19 898 and 28 055 bp. While palindromes II and III have 1 T-to-A substitution, they are located at 23 648 and 26 061 bp. A further BLAST analysis with the maxicircles of T. b. brucei (Lister 427, MN904526.1), T. b. equiperdum (STIB 818, EU185799.1), Trypanosoma congolense (IL3000, GCA_003013265.1) and Trypanosoma vivax (Y486, MT090068.1) enabled the identification of similar palindromes in these species (Fig. 5D), only 1 of each species is shown for illustrative purposes. These palindromes are highly conserved and contain an A5C element.
Unlike highly conserved coding regions, DRs show species specificity among trypanosomes (Fig. S5). It displays about 70% sequence identity in DRI between the T. musculi and T. lewisi maxicircle, while there are only some similar sequences (~400 bp) in DRII. Moreover, there are no other homologous sequences between T. musculi and the other 2 species (T. cruzi and T. brucei) in the DRs (Fig. S5). Therefore, these results suggest that DRII is highly divergent among trypanosomes, which may have the potential to be a good molecular marker for distinguishing T. musculi from related species.
Discussion
In this study, a sequence of the 34 606 bp kDNA maxicircle genome from T. musculi was reported and an in-depth investigation of T. musculi maxicircle sequences and comparative analysis with other Trypanosomatidae species were also undertaken. The size of the total coding region of the T. musculi kDNA maxicircle is 16 975 bp with 2 pronounced insertions in T. musculi MURF2 (630 bp), ND5 (1278 bp) and 1 deleted fragment of ND4 (281 bp) (Fig. 3A). It is different from T. brucei, T. cruzi and T. lewisi, in which their sizes are around 15 000 bp length. The 2 insertions in T. musculi maxicircle genes correspond either to a partial minicircle (630 bp) containing one of the CSBs or to a complete minicircle (1278 bp) containing 2 CSBs. Such insertions have not been observed in other Trypanosomatidae species except Leishmania donovani (1S LdBob strain) and T. cruzi (TcV strain) where the insertions were considered to be derived from minicircles due to CSBs. Therefore, the insertions were also thought to be a consequence of gene translocation, from minicircles to maxicircles (Nebohácová et al., Reference Nebohácová, Kim, Simpson and Maslov2009; Berná et al., Reference Berná, Greif, Pita, Faral-Tello, Díaz-Viraqué, Souza, Vallejo, Alvarez-Valin and Robello2021). Mostly gRNA genes are encoded in minicircles, but some gRNA genes, such as gMurf2 (30–79) and gNd7 (216–252) (Koslowsky et al., Reference Koslowsky, Sun, Hindenach, Theisen and Lucas2014; Li et al., Reference Li, Zhang, Lukeš, Li, Wang, Qu, Hide, Lai and Lun2020), were reported to be encoded in maxicircles in T. brucei and T. lewisi, respectively. Moreover, 7 maxicircle-encoded gRNAs were identical in L. tarentolae LEM125 and UC strains, which mediate the editing of Cyb, MURF2, A6 and ND7 transcripts (Simpson et al., Reference Simpson, Douglass, Lake, Pellegrini and Li2015). It can be assumed that the insertions deriving from minicircles may also possibly encode gRNA genes for RNA editing, therefore these may be an intermediate stage indicating that maxicircle encoding gRNA genes have originated from minicircles.
Maxicircle gene deletions are only rarely found in Trypanosomatidae species, such as similar deletions seen in ND4 of the T. cruzi Esmeraldo strain (Westenberger et al., Reference Westenberger, Cerqueira, El-Sayed, Zingales, Campbell and Sturm2006), and ND7 gene from asymptomatic T. cruzi isolates (Baptista et al., Reference Baptista, Vêncio, Abdala, Carranza, Westenberger, Silva, Pereira, Galvão, Gontijo, Chiari, Sturm and Zingales2006). The effect of these insertions and deletions on the parasite life cycle is still unclear. ND5 and ND4 are known as non-edited genes in other known Trypanosomatidae species and it is inconceivable that these large insertions/deletions could be corrected by U-insertion/deletion editing of the mRNAs. Nevertheless, all of the above insertions/deletions are found in ND4, ND5 as well as ND7 genes, and these genes all encode subunits of the mitochondrial respiratory chain NADH-dehydrogenase (Complex I). Since the presence of a functional Complex I in Trypanosomatidae species has long been debated (Opperdoes and Michels, Reference Opperdoes and Michels2008; Duarte and Tomás, Reference Duarte and Tomás2014). Deletions in kDNA encoding Complex I subunits were identified in some strains of T. cruzi that seem no impact in mitochondrial bioenergetics, ROS production or redox state in this parasite (César Carranza et al., Reference César Carranza, Kowaltowski, Mendonça, de Oliveira, Gadelha and Zingales2009). Although the presence of Complex I and its involvement in respiration has been clearly demonstrated in T. brucei, it appears to be non-essential for procyclic forms (Beattie and Howton, Reference Beattie and Howton1996; Verner et al., Reference Verner, Čermáková, Škodová, Kriegová, Horváth and Lukeš2011; Surve et al., Reference Surve, Heestand, Panicucci, Schnaufer and Parsons2012). The lack of editing in several Complex I subunits in L. tarentolae UC strain also suggests that it may not be essential (Simpson et al., Reference Simpson, Douglass, Lake, Pellegrini and Li2015). Therefore, the possibility that the role of Complex I subunits is less important in T. musculi was favoured with the presence of insertions/deletions in ND5 and ND4. In addition, another insertion occurs in MURF2, whose function remains uncertain but hypothesis could be risen. MURF2 might be a new component in Complex I. The insertion in MURF2 may be a recent event as it is only found in Partinico II strain, but not other 3 strains of T. musculi. The loss of conservation in MURF2 could probably be attributed to the loss of function of Complex I components and consequently on selection pressures on the gene. To verify this hypothesis, a highly sensitive and accurate identification of the functioning of Complex I in Trypanosomatidae species would be interesting to investigate.
The DR of the kinetoplast maxicircle was initially described as a variable and non-coding region and the DR structure seemed to be drastically different in various species (Borst et al., Reference Borst, Fase-Fowler, Hoeijmakers and Frasch1980, Reference Borst, Weijers and Brakenhoff1982; Stuart and Gelvin, Reference Stuart and Gelvin1982; Muhich et al., Reference Muhich, Simpson and Simpson1983; Maslov et al., Reference Maslov, Kolesnikov and Zaitseva1984). Therefore, the function of the DRs remains as an enigma. Studies on T. brucei, Crithidia oncopelti, Leptomonas collosoma and Leishmania seymouri revealed some CSBs-like sequences in their maxicircle DRs. As CSBs are essential for minicircle replication (Ryan et al., Reference Ryan, Shapiro, Rauch and Englund1988), CSBs-like sequences may play a similar role in maxicircle replication (Gorbat et al., Reference Gorbat, Maslov, Peters, Gaviernik, Viustenkhagen and Kolesnikov1990; Sloof et al., Reference Sloof, de Haan, Eier, van Iersel, Boel, van Steeg and Benne1992; Myler et al., Reference Myler, Glick, Feagin, Morales and Stuart1993; Flegontov et al., Reference Flegontov, Guo, Ren, Strelkova and Kolesnikov2006). However, CSB-I or III-like regions were not identified in T. musculi DRs, instead, only a CSB-II-like region (CCCGTGT) is located at 19 817 bp. CSB-I or III-like regions were also not found in DRs of the closely related T. lewisi, suggesting a CSB-independent maxicircle replication mechanism exists in these species. Therefore, although CSB-like sequences were present in the insertions of T. musculi MURF2 and ND5, it is not clear whether they are also involved in maxicircle replication.
It has been demonstrated that hairpins or cruciform structures (palindromes) are frequently associated with promoters and may also act as protein-binding sites (Wadkins, Reference Wadkins2000). Palindromes with an A5C-element in DRs are suggested as recognition sites for binding of transcription factors or transcription initiation (Vasil'eva et al., Reference Vasil'eva, Bessolitsina, Merzlyak and Kolesnikov2004; Flegontov et al., Reference Flegontov, Guo, Ren, Strelkova and Kolesnikov2006). Some palindromes also have been identified in T. musculi as well as in a variety of other Trypanosomatidae species, where each consists of 1 A5C-element. It may be speculated that these palindromes play a significant role in Trypanosomatidae species maxicircles, judged by their high degree of sequence conservation in the evolution of Kinetoplastida species.
The trypanosome maxicircle presents itself as a complex evolutionary system, and it may be an excellent taxonomic marker in phylogenetic analysis. The coding region of the maxicircle in phylogenetic analyses provides a robust evolutionary insight into the relationships within Trypanosomatidae species (Lin et al., Reference Lin, Lai, Zheng, Wu, Lukeš, Hide and Lun2015; Kaufer et al., Reference Kaufer, Stark and Ellis2019; Kay et al., Reference Kay, Williams and Gibson2020). A close affinity between T. musculi and T. lewisi in Herpetosoma was also supported, which clustered with the sister groups of Schizotrypanum and Aneza. Unlike the highly conserved coding region, the DRs of maxicircle, especially DRII sequence, was found to be significantly divergent and species-specific (Kay et al., Reference Kay, Williams and Gibson2020). The homologies in DRII between closely related species, e.g., T. musculi and T. lewisi, phylogenetic clades of T. cruzi, are limited (Figs S5 and S8). Such a characteristic of DRII provides an opportunity for developing a valuable molecular marker for distinguishing closely related species and subspecies. Actually, a preliminary test on 3 T. musculi strains and 6 T. lewisi strains revealed a consistent amplification of DRII fragments, which could enable them to be distinguished from each other and 13 strains of other trypanosomes (Hong et al., Reference Hong, Zhang, Fusco, Lan, Lun and Lai2017).
In general, this study reports the first detailed description and analysis of the kDNA maxicircle genome of T. musculi and reveals a relatively high overall conservation of gene content and synteny with other trypanosome species. Furthermore, the divergence of DRII suggests its potential as a valuable marker for distinguishing these evolutionarily related species.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022001019
Acknowledgements
The authors would like to thank Dr Ling-Ling Zheng for help with bioinformatics analysis. Thanks to all the members in the laboratories who provided useful help during the field and laboratory work. We would also like to appreciate the critical comments from the anonymous reviewers which have greatly helped to improve our paper.
Author's contributions
J-F Wang, R-H Lin, D-H Lai and Z-R Lun designed the study. J-F Wang, R-H Lin and X Zhang conducted data gathering and performed statistical analyses. J-F Wang, R-H Lin and D-H Lai drafted the manuscript and undertook data extraction and screening. G Hide, ZR Lun and DH Lai critically reviewed the paper. All authors approved the final version and agree to be accountable for all aspects of the work.
Financial support
The project was supported by grants from the National Natural Science Foundation of China (31672276, 31720103918) and the Natural Sciences Foundation of Guangdong Province (2022A1515011874).
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical standards
The animal ethical approval of sample collection was obtained from the Institutional Review Board of Animal Care at Sun Yat-Sen University (License no. 31672276).
Data availability
Nucleotide sequence data reported in this paper are available in GenBank databases under accession numbers: Trypanosoma musculi maxicircle sequence (OM000218), Trypanosoma lewisi maxicircle sequence (OM000219), Trypanosoma grayi maxicircle sequence (OM049542), and Trypanosoma brucei rhodesiense maxicircle sequence (OM049543). PacBio and Illumina sequencing data have been deposited in NCBI's Sequence Read Archive (SRA) with BioProject ID PRJNA792722.