Introduction
In the last years, cryptic parasite species were continuously discovered in various helminth groups (Pérez-Ponce de León and Poulin, Reference Pérez-Ponce de León and Poulin2017). This can mainly be attributed to the application of molecular techniques that often provide a higher taxonomic resolution than morphological characters. The same trend was also found within the Acanthocephala, with several cryptic species discovered in various genera (e.g. Steinauer et al. Reference Steinauer, Nickol and Ortí2007; Wayland et al. Reference Wayland, Vainio, Gibson, Herniou, Littlewood and Väinölä2015). Acanthocephalans have a complex life cycle, including at least one obligatory host change. Arthropods, such as crustaceans or insects, serve as intermediate hosts whereas fish, birds, mammals and amphibians are known as final hosts, where reproduction occurs in the intestine. In some species, additional paratenic hosts can exist (Taraschewski, Reference Taraschewski2000; Kennedy, Reference Kennedy2006; Sures, Reference Sures and Schmidt-Rhaesa2014). Acanthocephalans belonging to the genus Polymorphus have an aquatic life cycle with waterfowl as final hosts in most species. In their amphipod intermediate hosts, the parasites migrate into the haemocoel where they develop to the cystacanth stage. The final host gets infected by ingesting the infected amphipod (Taraschewski, Reference Taraschewski2000; Kennedy, Reference Kennedy2006). The life cycles of some species of the Polymorphidae have not been fully understood to date and molecular barcoding has been applied successfully to link parasite stages in the intermediate and definitive hosts (Alcántar-Escalera et al. Reference Alcántar-Escalera, García-Varela, Vázquez-Domínguez and Pérez-Ponce de León2013).
The species Polymorphus minutus was originally described in Germany by Goeze (Reference Goeze1782) and later found in a wide variety of aquatic birds (e.g. Lühe, Reference Lühe and Brauer1911). Since the elucidation of the complete life cycle of P. minutus, various species of freshwater amphipods were identified as intermediate hosts (Greef, Reference Greef1864; Schmidt, Reference Schmidt, Crompton and Nickol1985). An experimental transmission study with P. minutus from the 1950s indicated that there is a possible differentiation in lineages or even cryptic species displaying different intermediate host specificities (Hynes and Nicholas, Reference Hynes and Nicholas1958): P. minutus cystacanths isolated from Gammarus duebeni, Gammarus pulex and Gammarus lacustris were used to infect domestic ducks, respectively. The resulting eggs obtained from the adult parasites were used to infect each of the three Gammarus spp. The results clearly showed that the eggs were most infective for the same Gammarus species from which the respective cystacanths originated. Despite these observations, it was commonly assumed that specimens of Polymorphus collected in Central Europe all belong to a single species. Accordingly, no recent taxonomic studies were published for this species. Furthermore, P. minutus has been frequently used as a model organism in various fields throughout the last decades, including studies about tegumental structure of adults (Crompton and Lee, Reference Crompton and Lee1965) and acanthor-larvae (Albrecht et al. Reference Albrecht, Ehlers and Taraschewski1997) or the relation of the parasite with its intermediate and final hosts (Hynes and Nicholas, Reference Hynes and Nicholas1963; Itämies et al. Reference Itämies, Valtonen and Fagerholm1980; Bollache et al. Reference Bollache, Gambade and Cézilly2001; Bauer et al. Reference Bauer, Haine, Perrot-Minnot and Rigaud2005; Haine et al. Reference Haine, Boucansaud and Rigaud2005; Médoc et al. Reference Médoc, Bollache and Beisel2006; Tain et al. Reference Tain, Perrot-Minnot and Cézilly2006; Jacquin et al. Reference Jacquin, Mori, Pause, Steffen and Medoc2014). If the hypothesis holds true that several cryptic species are subsumed in a presumed ‘P. minutus-complex’, this will have consequences for the conclusions drawn from behavioural or structural studies, especially if different intermediate host species were involved. Therefore, it is particularly essential to resolve the taxonomy of Polymorphus cf. minutus, also bearing in mind that this species can lead to high mortalities in waterfowl populations and is recognized as an economically significant pest in goose and duck farming (Hynes and Nicholas, Reference Hynes and Nicholas1963; Itämies et al. Reference Itämies, Valtonen and Fagerholm1980). However, the morphological differentiation of Polymorphus species is difficult, especially based on the cystacanth stage (Alcántar-Escalera et al. Reference Alcántar-Escalera, García-Varela, Vázquez-Domínguez and Pérez-Ponce de León2013). Furthermore, closely related parasite species might not provide defined and easily visible morphological characters for an unambiguous species diagnosis (Nadler and Pérez-Ponce de Léon, Reference Nadler and Pérez-Ponce de León2011; Selbach et al. Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015). Therefore, we used molecular data – mitochondrial cytochrome c oxidase subunit 1 (COI) fragment and the nuclear internal transcribed spacer (ITS)1-5.8S-ITS2 region – to identify and distinguish the Polymorphus-isolates sampled from amphipod populations in Germany and France. The aim was to assess the genetic diversity of what is considered P. minutus and its linkage to different intermediate amphipod hosts, thereby testing previous results from transmission experiments which pointed to a so far neglected cryptic species diversity (Hynes and Nicholas, Reference Hynes and Nicholas1958).
Materials and methods
Amphipod and parasite sampling
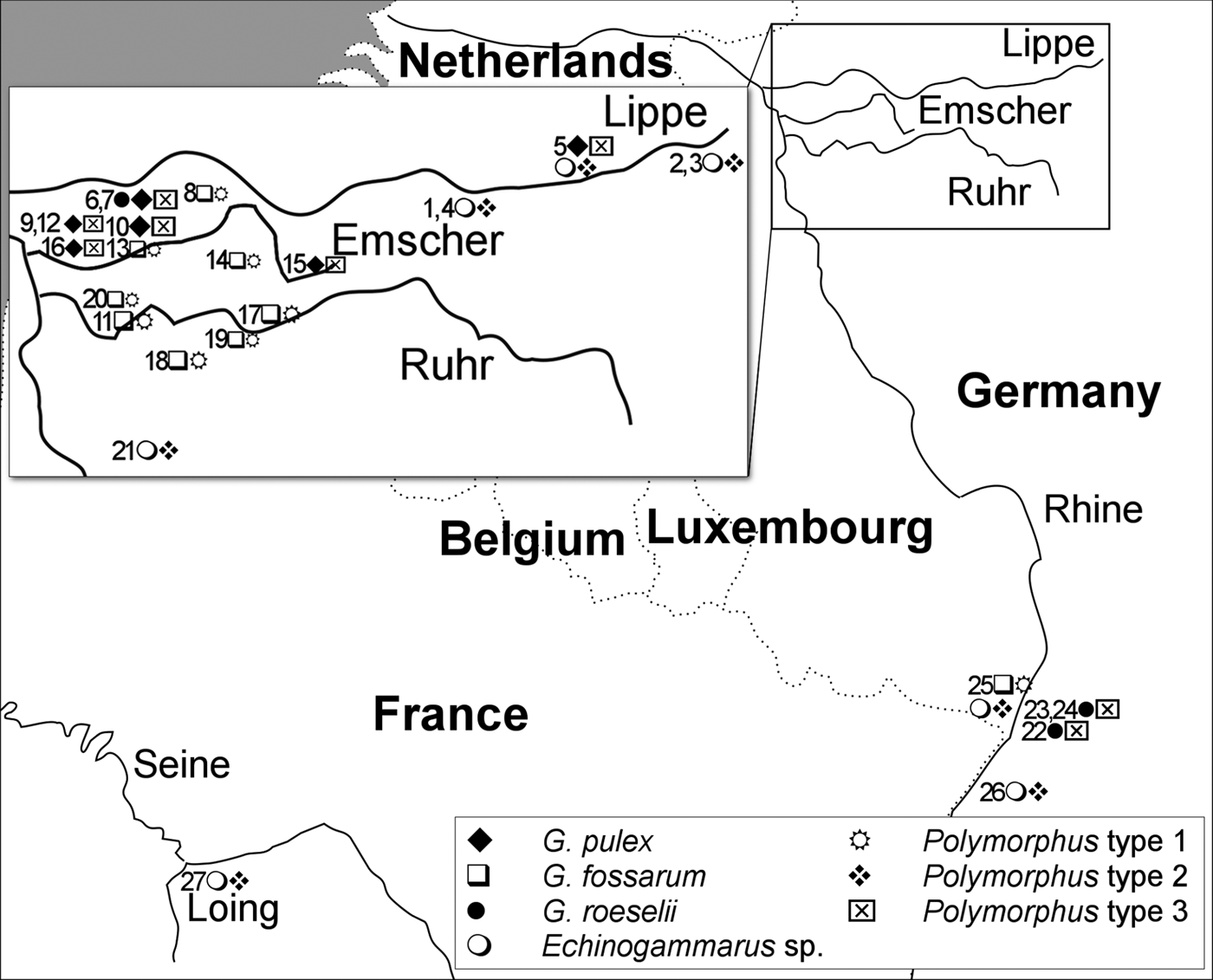
Gammarids infected with acanthocephalan cystacanths were sampled by kick-sampling, using a net with a mesh size of 0.3 mm, between 2014 and 2016 at 27 sites in North-Rhine Westphalia, Germany (smaller tributaries of Lippe, Emscher, Ruhr and Rhine), Baden-Wuerttemberg and Rhineland-Palatinate, Germany (tributaries of the Rhine), as well as Île-de-France, France (Loing, tributary of Seine River) (see map in Fig. 1). A list of all sites, sampling dates and the number of sequenced host individuals and Polymorphus cystacanths, as well as the geographic location, is given in Supplementary Table S1.

Fig. 1. Map of sampling area. The map shows the major river catchments, the approximate location of the sampling sites with the amphipod and Polymorphus types found. Sites 1–8: Lippe (Rhine), 9–16: Emscher (Rhine), 17–20: Ruhr (Rhine), 21–26: small tributaries of the Rhine, 27: Loing (Seine).
Infected individuals were sorted at the site according to the obvious red coloration of the cystacanths and stored in 95% ethanol. In the laboratory, amphipods were identified morphologically (i.e. for. Gammarus roeselii and Echinogammarus sp.) according to the taxonomic keys of Eggers and Martens (Reference Eggers and Martens2001, Reference Eggers and Martens2004). Due to morphological difficulties to identify species of the G. pulex and G. fossarum cryptic species complexes, those were DNA barcoded. The material of the host was not retained in all cases (e.g. when additional collection material was included which had been collected before the study was designed), therefore, a genetic identification of hosts was not always possible (see Supplementary Table S1).
PCR and sequencing
Cystacanths were dissected from gammarids and DNA extraction of hosts was performed according to Grabner et al. (Reference Grabner, Weigand, Leese, Winking, Hering, Tollrian and Sures2015). DNA from small pieces of the cystacanths was extracted with the same protocol. The standard animal barcoding locus COI was amplified for amphipods and acanthocephalans using the degenerated primer pair LCO1490-JJ (5′-CHA CWA AYC ATA AAG ATA TYG G-3′) and HCO2198-JJ (5′-AWA CTT CVG GRT GVC CAA ARA ATC A-3′) of Astrin and Stüben (Reference Astrin and Stüben2008). Each PCR reaction mix (total volume of 12.5 µL) contained 1 µL template DNA, 0.2 mm dNTPs, 1 × PCR buffer, 0.5 µ m of each primer, 0.025 U µL−1 Hotmaster Taq-polymerase (5 PRIME GmbH, Hamburg, Germany) and the rest water. PCR cycle conditions were as follows: initial denaturation for 1 min at 94 °C, followed by 36 cycles of 2 min at 94 °C (denaturation), 20 s at 50 °C (annealing) and 30 s at 65 °C (elongation), and a final elongation step for 5 min at 65 °C.
For some of the acanthocephalans, the nuclear ITS1-5.8S-ITS2 loci were additionally investigated to validate COI results using a newly developed Polymorphus-specific primer pair pmF (5′-CCT CAC GGT AAT TCT ATC AG TC-3′) and pmR (5′-CGC AAT CGT GTC ATC TCA GT-3′). The same PCR reaction mix volumina were used, and the following PCR cycle conditions applied: initial denaturation for 5 min at 94 °C, followed by 37 cycles of 40 s at 94 °C (denaturation), 35 s at 56 °C (annealing) and 35 s at 68 °C (elongation), and a final elongation step for 5 min at 68 °C.
Prior to the sequencing of the COI and the nuclear fragment, an ExoI/FastAP purification step was performed. For this purpose, 9 µL of each PCR product was mixed with 1 µL FastAP (1 U µL−1) and 0.5 µL ExoI (20 U µL−1), both from Thermo Fisher Scientific (Schwerte, Germany). The PCR products were enzymatically purified at 37 °C for 25 min and at 85 °C for 15 min. The PCR products were sequenced at GATC Biotech AG (Köln, Germany) using the respective PCR primer pair.
Genetic analyses
Three separate alignments of the resulting host (COI) and parasite sequences (COI and ITS1) were created in Geneious v.6.0.5 (Kearse et al. Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Mentjies and Drummond2012) using the Muscle plug-in with three iterations. Alignments were trimmed by first removing the primer sequences and second using the GBLOCKS web server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html), thereby further accounting for and automatically removing poorly aligned flanking regions (e.g. resulting from ambiguities or missing data). Final alignments had a length of 658 bp (COI hosts), 432 bp (COI parasites) and 311 bp (ncDNA loci), with the latter encompassing the 3′-region of the ITS1 locus, the complete 5.8S rRNA gene (162 nucleotides) and a minor fragment of the 5′-end of the ITS2 locus (25 nucleotides, excluded from downstream analyses) as identified by comparison with the annotated ITS1-5.8S-ITS2 P. minutus sequence available in NCBI GenBank (AY532067).
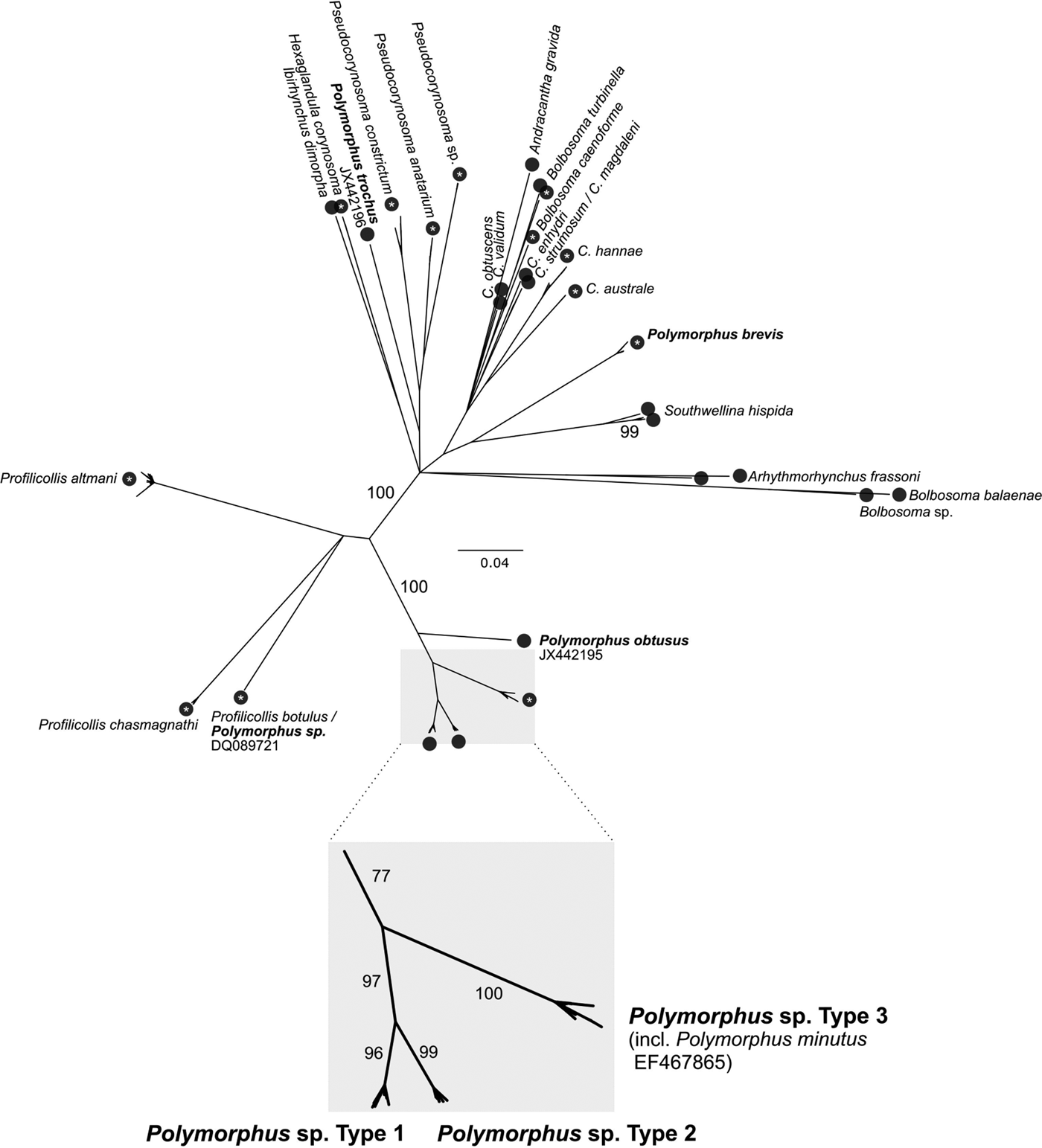
The number of potential cryptic Polymorphus cf. minutus species was calculated by the Automatic Barcode Gap Discovery (ABGD) method (Puillandre et al. Reference Puillandre, Lambert, Brouillet and Achaz2012). For this purpose, all available COI sequences (excluding pseudogenes) with a length >500 bp were retrieved from GenBank as on 08.08.2017 (n = 322). Sequences were aligned using the Muscle-plugin of Geneious (three iterations) and trimmed to the 655 bp (for acanthocephalans) and 658 bp (for gammarids) long Folmer-fragment. The Polymorphidae COI alignment was combined with the COI parasite alignment of this study. Identical sequences were deleted using the web application ElimDupes (https://hcv.lanl.gov/content/sequence/ELIMDUPES/elimdupes.html), resulting in 264 unique Polymorphidae sequences from NCBI and 126 from this study. Because sequences had unequal lengths, the COI consensus alignment (n = 390) was further trimmed using the GBLOCKS web server to a total alignment length of 423 bp. The ABGD method was performed on this alignment using the following final setting: Pmin 0.001; Pmax 0.1; Steps: 50; using a Kimura K80 model (ratio 1.5). A Neighbour-Joining (NJ) tree was constructed for visualization of the ABGD clusters using the Geneious Tree Builder (tree building method: NJ; genetic distance model: Jukes–Cantor; 500 bootstrap replicates).
Amphipod hosts were identified by comparing the obtained COI sequences with the Barcode of Life Datasystem reference library (BOLD; Ratnasingham and Hebert, Reference Ratnasingham and Hebert2007).
Parasite COI sequences of length <450 bp were used for identification purpose only and excluded from subsequent intraspecific genetic diversity analyses. Haplotype networks were calculated in PopArt (Leigh and Bryant, Reference Leigh and Bryant2015) using the TCS option (Clement et al. Reference Clement, Posada and Crandall2000). Haplotype and nucleotide COI diversity were calculated in DnaSP v5 (Librado and Rozas, Reference Librado and Rozas2009). Intraspecific genetic distances were calculated with MEGA6 (Tamura et al. Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). The ITS1-5.8S alignment was used for visual identification of nuclear alleles.
Results
Amphipod and parasite sampling
At each site, a variable number of one to 33 Polymorphus-infected amphipods was available for genetic analyses, largely depending on local prevalences (see Supplementary Table S1). Sequencing of the intermediate hosts confirmed the morphological identification, except cases when no amphipod samples were available for molecular analyses. At sites 25 and 26, Echinogammarus specimens were not identified to species level and are treated as Echinogammarus sp. (either E. ischnus or E. trichiatus, the only described Echinogammarus spp. in Germany besides E. berilloni that can be identified easily). Polymorphus infections were found in at least five amphipod species (Echinogammarus berilloni, Echinogammarus sp., Gammarus fossarum type B, G. pulex, G. roeselii). At two locations, more than one Polymorphus-infected amphipod species were present: site 5 (Lippe, G. roeselii and E. berilloni) and site 25 (Lauter, Echinogammarus sp. and G. fossarum). Usually, intermediate hosts were infected by a single cystacanth. However, two and three Polymorphus individuals occurred in single intermediate host specimens at four sites: site 4 (three cystacanths in E. berilloni), site 11 (three in G. fossarum), site 17 (2 × two in G. fossarum) and site 20 (3 × two in G. fossarum). At five sites (sites 1, 2, 22, 25, 27), amphipod populations were additionally infected by the acanthocephalan parasite Pomphorhynchus laevis (see Supplementary Table S1 for details).
Interspecific genetic diversity of the parasite
All individual COI (n = 145), ITS1 (66) and 5.8S (66) parasite sequences were deposited in the BOLD project ‘PACDE’ with BOLD identifiers PACDE001-18 to PACDE178-18. Sequencing of the parasite COI locus revealed the presence of three cryptic Polymorphus lineages, as of now referred to as Polymorphus type 1-3 (PspT1-3). Molecular species delimitation via ABGD resulted in 30 clusters for the investigated COI sequences of the full Polymorphidae dataset retrieved from GenBank, providing further support for the three Polymorphus types (Fig. 2). Among those, specimens of PspT3 demonstrated a maximum sequence identity of 99% to the COI (GenBank accession no. EF467865.1) and ITS sequence (AY532067.1) of P. minutus available in GenBank, respectively (originating from G. pulex from Dijon, France as published in García-Varela and Pérez-Ponce de León, Reference García-Varela and Pérez-Ponce de León2008 and García-Varela et al. Reference García-Varela, Aznar, Pérez-Ponce de León, Piñero and Laclette2005). PspT1 was most similar (99%) to JF803287.1 in GenBank (from G. fossarum in Switzerland, published in Westram et al. Reference Westram, Baumgartner, Keller and Jokela2011).

Fig. 2. Molecular Species Delimitation. COI Neighbour-Joining tree for visualization of molecular species delimitation results. Black circles represent clusters proposed by the ABGD method. White asterisks within the circles indicate a 100% bootstrap support for the respective cluster. Further bootstrap supports are provided at the branches.
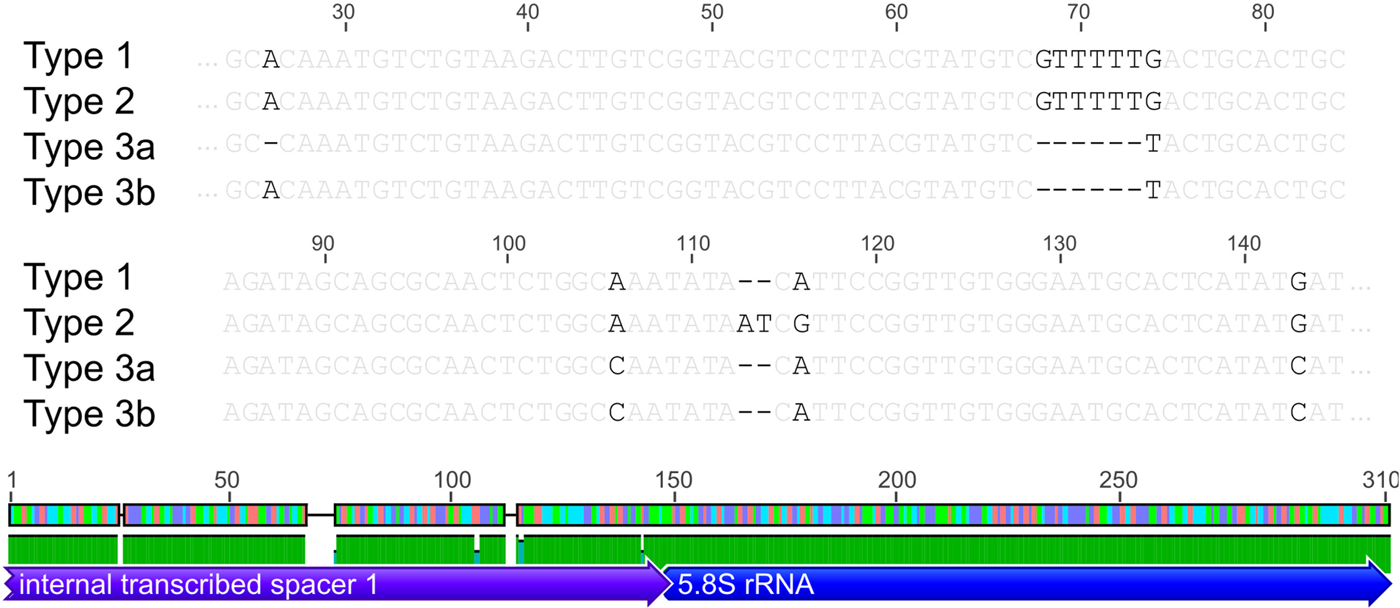
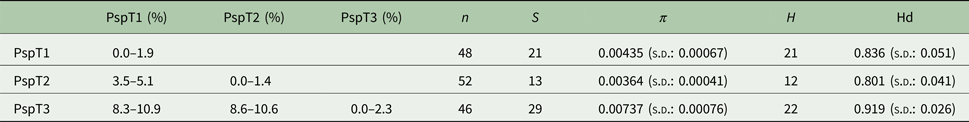
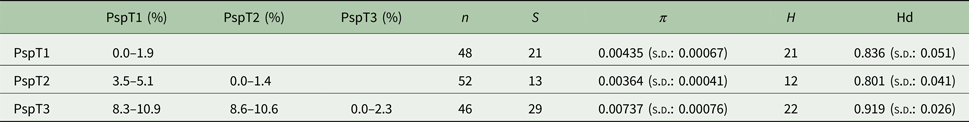
Based on the investigated COI locus, the three Polymorphus types are genetically differentiated by at least 3.5% (PspT1/PspT2), with a maximum interspecific distance of 10.9% between specimens of PspT1 and PspT3 (Table 1). The pattern revealed by the nuclear ITS1 marker is in full congruence with the three mitochondrial COI lineages observed: PspT1 and PspT2 both have one specific ITS1 allele each, with only PspT3 possessing two but likewise specific ITS1 alleles, which are differentiated by one mutation (Fig. 3). The 5.8S rDNA was completely identical for all investigated specimens. For 35 specimens (8 × PspT1, 6 × PspT2, 21 × PspT3), we obtained both the mitochondrial COI and the nuclear ITS1-5.8S fragment, linking the different datasets (Supplementary Table S1 and Supplementary Fig. S1).
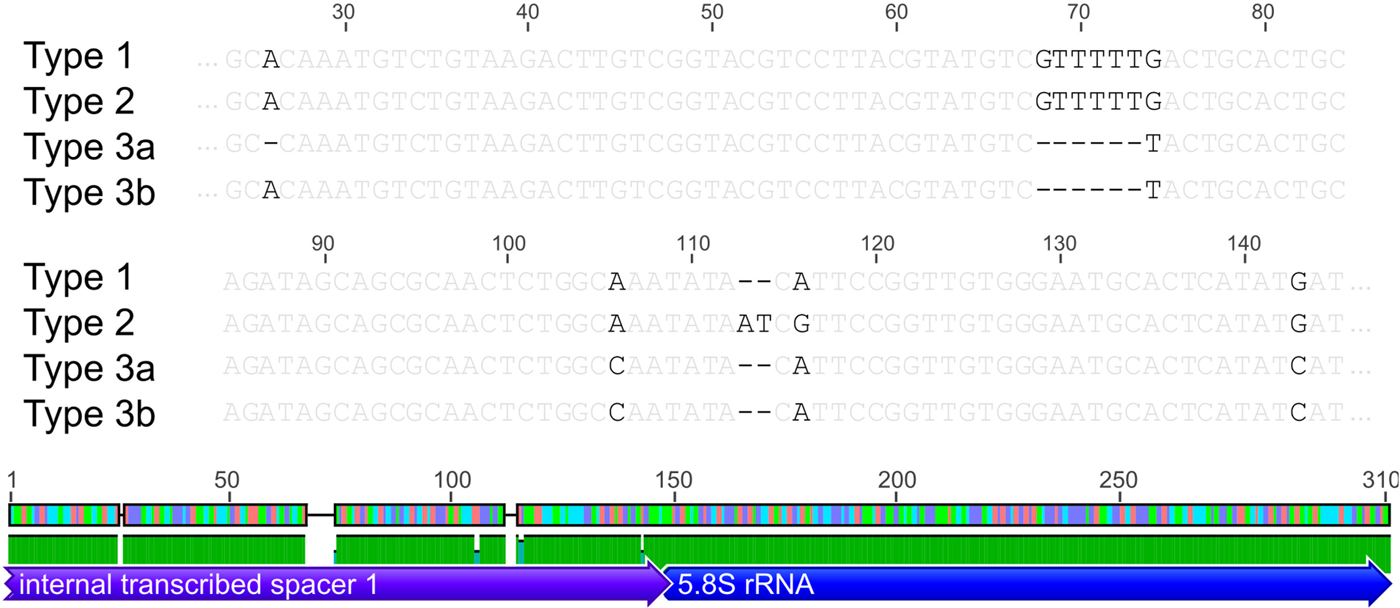
Fig. 3. Nuclear ITS1 alleles. Indication of nuclear ITS1 alleles for the three Polymorphus sp. types. Base pair differences are highlighted in black. Polymorphus sp. types 1 and 2 possess one ITS1 allele each, whereas Polymorphus sp. type 3 demonstrates two, distinguished by one mutation. The 5.8S rDNA was completely identical for all investigated specimens. Figure elements are partly used from Geneious v.6.0.5 (Kearse et al. Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Mentjies and Drummond2012).
Table 1. Genetic distances, nucleotide and haplotype diversity based on the COI fragment of the Polymorphus types
Intraspecific genetic diversity
Maximum values of intraspecific genetic distances vary from 1.4% in PspT2 to 2.3% in PspT3 (Table 1). Nucleotide diversity for all three Polymorphus types is moderate (π: 0.004–0.007), whereas haplotype diversity is very high (hd: 0.80–0.92). The highest number of haplotypes is observed in PspT3 (H = 22), followed by PspT2 (21) and PspT1 (12). The most frequently observed haplotypes are centred within the haplotype networks, which show characteristic star-like patterns (Fig. 4). Since the number of investigated parasite specimens for each Polymorphus type were inconsistent for the different localities, only specimen-based haplotype diversity is depicted and no further site-specific analyses were performed (Supplementary Table S1).

Fig. 4. COI haplotype networks for the three Polymorphus sp. types are indicated. The size of the circles is proportional to the frequency of the respective haplotype encountered in each of the three datasets. Grey dots represent hypothetical haplotypes not sampled within this study. Lines between (hypothetical) haplotypes indicate one mutational step. Spatial data for all haplotypes are given (NRW: North Rhine-Westphalia; BW: Baden-Württemberg; F: France). For PspT3, the distribution of the individual haplotypes in the two hosts, G. pulex (Gp) and G. roeselii (Gr) is indicated. The arrow points to a Polymorphus sp. cystacanth in its amphipod intermediate host.
Intermediate host specificity
The three Polymorphus types are widely distributed and demonstrate a high level of intermediate host specificity (Fig. 1). PspT1 was exclusively found in G. fossarum type B (10 sites). PspT2 was only found in Echinogammarus sp./E. berilloni (eight sites). PspT3 was restricted to G. pulex (six sites) and G. roeselii (five sites). At sites where two Polymorphus types were co-occurring, each of the two types was found only in a single intermediate host species: site 5 (Klostermersch, PspT2 in E. berilloni and PspT3 in G. roeselii) and site 25 (Lauter, PspT1 in G. fossarum and PspT2 in Echinogammarus sp.).
Discussion
In the present study, we investigated the genetic diversity of the acanthocephalan endoparasite P. minutus (Goeze, Reference Goeze1782). Three cryptic species were detected (Polymorphus type 1–3; PspT1-3), which are congruently supported by mitochondrial (COI) and nuclear (ITS1) data. Our results further indicate that all three potential species are specific for their intermediate amphipod host genus or species.
Cryptic species are a common phenomenon among the Acanthocephala (Wayland, Reference Wayland2010; Pérez-Ponce de León and Poulin, Reference Pérez-Ponce de León and Poulin2017). Even for our target organism, P. minutus, there have been indications of different intermediate host specificities (Hynes and Nicholas, Reference Hynes and Nicholas1958) that might be attributable to cryptic Polymorphus species. Recent results also indicate morphological differences between isolates of P. minutus (Zittel and Taraschewski, unpublished results); therefore the PspT1-3 might turn out to be pseudo-cryptic. However, until now, P. minutus was considered the only species of its genus in Central Europe frequently parasitizing amphipod intermediate hosts that are commonly used for ecotoxicological (summarized in Sures et al. Reference Sures, Nachev, Selbach and Marcogliese2017) and behavioral studies (see, e.g., Helluy, Reference Helluy2013 and references therein). In the light of our results of three cryptic and potentially intermediate-host specific Polymorphus cf. minutus lineages, the comparison of findings between studies based on different intermediate host–parasite systems has to be treated with caution. However, the most frequently investigated amphipods studied for the influence of parasite infections – G. roeselii and G. pulex (e.g. Bauer et al. Reference Bauer, Haine, Perrot-Minnot and Rigaud2005; Médoc et al. Reference Médoc, Bollache and Beisel2006; Lagrue et al. Reference Lagrue, Güvenatam and Bollache2013) – host the same parasite species (i.e. PspT3) and previous results are thus likely to be comparable.
For the study of parasite–host specificity, the correct identification of the host is crucial. Yet, morphological differentiation of G. pulex and G. fossarum specimens is particularly challenging, especially when no fully mature, adult individuals are available (Karaman and Pinkster, Reference Karaman and Pinkster1977). Additionally, Chen et al. (Reference Chen, Grabner, Nachev, Shih and Sures2015) have previously shown that infected individuals are often smaller and therefore more difficult to identify. Adding even more complexity, G. fossarum as well as G. pulex morphospecies are both known to comprise a high degree of cryptic lineages/species, sometimes showing even different ecological requirements (Lagrue et al. Reference Lagrue, Wattier, Galipaud, Gauthey, Rullmann, Dubreuil, Rigaud and Bollache2014; Weiss et al. Reference Weiss, Macher, Seefeldt and Leese2014; Grabner et al. Reference Grabner, Weigand, Leese, Winking, Hering, Tollrian and Sures2015; Eisenring et al. Reference Eisenring, Altermatt, Westram and Jokela2016). DNA barcoding of intermediate hosts and parasites may help to circumvent potential future misinterpretations based on misidentified host–parasite relationships, as both ‘cryptic dimensions’ have to be correctly assessed.
Nearly one-third of all infected intermediate host specimens detected in this study belonged to G. fossarum type B, being solely infected by PspT1, indicating a specific parasite–host relationship. This assumption is supported by a previous study on Polymorphus infections of Gammarus spp. populations in Switzerland (Westram et al. Reference Westram, Baumgartner, Keller and Jokela2011). In the latter study, a total of 58 populations have been investigated with 13 populations being infected by Polymorphus cf. minutus. Comparing the deposited ITS1 sequence from Westram et al. (Reference Westram, Baumgartner, Keller and Jokela2011) to our data, we can identify the Swiss Polymorphus lineage as PspT1 (with 99% sequence identity to JF803287). Only a single specimen of G. fossarum type A, but to the large majority G. fossarum type B were infected by PspT1, although eight G. pulex populations were investigated in the same study, of which two occurred in sympatry with PspT1-infected G. fossarum type B populations (see supplementary material of Westram et al. Reference Westram, Baumgartner, Keller and Jokela2011). The consistent finding of the same intermediate host–parasite relationship (i.e. PspT1 exclusively infecting G. fossarum type B) for another geographical region further strengthens the assumption of intermediate host specificity of PspT1. In the present study, PspT2 was detected exclusively in Echinogammarus spp. both from Seine and Rhine tributaries, which suggests that this is at least a genus-specific parasite–host association. PspT3 also seems to be less restricted to a single host species, as it was found in both G. pulex and G. roeselii, which is in accordance with the hypothesis of Kennedy (Reference Kennedy2006) that certain acanthocephalan species evolve with multiple intermediate host species.
Besides the results presented here, previous studies provide further evidence for the potential intermediate host specificity of the different Polymorphus types. Jacquin et al. (Reference Jacquin, Mori, Pause, Steffen and Medoc2014) detected Polymorphus sp. infections in E. berilloni, but not in the co-occurring G. pulex. In a study conducted in Ireland, Polymorphus sp. was detected in native G. duebeni celticus (in 2/70 and 9/65 specimens, respectively), but not in co-occurring non-native G. tigrinus (43 investigated specimens) and G. pulex (30 specimens) from the same sites (Dunn and Dick, Reference Dunn and Dick1998), taking into account that the number of specimens might have been too low to detect low prevalence infections.
Besides the amphipod species mentioned above, Polymorphus infections have also been observed in Echinogammarus stammeri (Dezfuli and Giari, Reference Dezfuli and Giari1999), Echinogammarus tibaldii (Dezfuli et al. Reference Dezfuli, Lui, Giovinazzo and Giari2008), Gammarus lacustris (Hynes and Nicholas, Reference Hynes and Nicholas1958), Gammarus oceanicus, Gammarus salinus and Gammarus zaddachi (Lehtonen and Hario, Reference Lehtonen and Hario1996) in different European countries. This diversity of intermediate hosts suggests that even more species within the Polymorphus cf. minutus complex might exist than those discovered in the present study.
The reason for intermediate host specificity in acanthocephalans might be seen in the adaptation to the invertebrate host immune response. For example, Hynes and Nicholas (Reference Hynes and Nicholas1958) found large numbers of dead larvae of P. minutus at the outer surface of the gut of experimentally infected amphipods when eggs of parasites were used that originated from cystacanths isolated from another amphipod species. This is in line with findings for other acanthocephalans showing that the migration of the parasite through the gut wall of the intermediate host in the early phase of infection is most crucial for survival (Taraschewski, Reference Taraschewski2000).
Based on our data, we can only speculate on the specificity of the three Polymorphus sp. types for their definitive hosts. Numerous bird species were listed as hosts for P. minutus (e.g. Lühe, Reference Lühe and Brauer1911), while anatid ducks and goose are probably the most common definitive hosts. Furthermore, there are some reports of Polymorphus spp. from water-related rodents such as water shrews or muskrats (summarized in Platt, Reference Platt1978). The question, if the three Polymorphus types identified in the present study show some sort of definitive host specificity is highly relevant for the understanding of the biology of this genus. Either each of the respective Polymorphus-types will develop only in a specific definitive host species (due to species-specific strategies overcoming the hosts immune systems), or if they colonize the same definitive host individual, some mechanism of reproductive isolation must exist (e.g. by preferring different sections of the host gut, or by showing different seasonality patterns). Due to a lack of Polymorphus-specimens from definitive hosts, we can only speculate here. Also, we are not able to trace back which Polymorphus type actually refers to P. minutus (Goeze, Reference Goeze1782), initially described as Echinorhynchus minutus, potentially from Melanitta fusca and Turdus merula (pages 164 & 165 in Goeze, Reference Goeze1782) in Germany. It is questionable if this will ever be possible due to the lack of morphological characters given in these early studies (Smales, Reference Smales and Schmidt-Rhaesa2014).
The present study also provides evidence for at least one non-native acanthocephalan in Germany: PspT2 was detected only in Echinogammarus, most of which were E. berilloni. This amphipod is an established, non-native species in Germany that originated from the Mediterranean region (France and Spain) and that was recorded for the first time in Germany in 1924 (Tittizer et al. Reference Tittizer, Schöll, Banning, Haybach and Schleuter2000). As PspT2 was not found in native amphipods, it seems likely that E. berilloni introduced this Polymorphus type to Central Europe. Also, PspT3 might be a non-native parasite that was brought to Central Europe a few hundred years ago by the invasion of G. roeselii from the Balkan region (Grabowski et al. Reference Grabowski, Mamos, Bącela-Spychalska, Rewicz and Wattier2017). If this scenario is true, PspT3 must have colonized G. pulex as an additional host species. Nevertheless, it seems more likely that G. pulex is the native host for PspT3 and G. roeselii was colonized later, given that PspT3: (a) occurs in an indigenous amphipod species (G. pulex) and (b) is the genetically most diverse parasite lineage in our dataset (in terms of nucleotide and haplotype diversity), not pointing to any severe decline in genetic diversity due to potential historical bottleneck events during invasion processes. Recently, evidence for another non-native acanthocephalan in the Rhine system was presented (Hohenadler et al. Reference Hohenadler, Nachev, Thielen, Taraschewski, Grabner and Sures2017). In the case reported by the latter authors, the fish infecting acanthocephalan Pomphorhynchus laevis was most likely introduced by invasive goby species that can be used as paratenic hosts for this parasite. In addition, the eoacanthocephalan Paratenuisentis ambiguus is also a non-native species (Taraschewski et al. Reference Taraschewski, Moravec, Lamah and Anders1987), originating from North America where it was described from Gammarus tigrinus as an intermediate host and the American eel as the definitive host. Interestingly, following the transplantation of G. tigrinus into the rivers Weser and Werra in the late 1950s, this acanthocephalan started to migrate into different rivers and accepted the European eel as a new definitive host, where it reached high prevalences and intensities (Sures and Streit, Reference Sures and Streit2001). In contrast to its final host switch, P. ambiguus was never described from other gammarid species than G. tigrinus, which also suggests a clear intermediate host specificity. Taken together, these examples show that recent species introductions are common for acanthocephalans, but the mechanisms of invasion vary.
In conclusion, we provide consistent support for the existence of at least three cryptic parasite species within Polymorphus cf. minutus, which very likely demonstrate a high degree of host specificity for their amphipod intermediate hosts. The immunological adaptation of the acanthocephalan to the intermediate host and the biochemical alterations caused by the parasite (Taraschewski, Reference Taraschewski2000; Helluy, Reference Helluy2013) might be a possible driver of this diversification. To correctly assess (intermediate) host–parasite relationships in the future, the application of routine DNA barcoding is highly encouraged (but see Pérez-Ponce de León and Nadler, Reference Pérez-Ponce de León and Nadler2010; Alcántar-Escalera et al. Reference Alcántar-Escalera, García-Varela, Vázquez-Domínguez and Pérez-Ponce de León2013). According to our results, we also suggest reviewing the taxonomy of Polymorphus spp. in Europe including adult stages to further clarify, whether the three Polymorphus types identified in the present study are valid species also differing in definitive host specificity and morphology.
Acknowledgements
The authors thank Jessica Schwelm for providing the Amphipod samples from the Klostermersch site and Dana Shilton from the Lippe River. Amphipods sampled at the Rhine tributaries (Alb, Lauter, Federbach, Knielinger See) were kindly provided by Anja Rüdiger, Anna-Lena Croneiß, Hans Kohls, Jochen Schwantzer and Thomas Walter. This publication is part of the EU COST Action DNAqua-Net (CA15219).
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182018000173.
Financial support
The Mercator Research Center Ruhr (MERCUR) is gratefully acknowledged for funding this study (Grant Pr-2014-0007).