Introduction
Monogenoidea Bychowsky, 1937 is a group of obligate parasites commonly found in freshwater and marine fishes (Boeger and Vianna, Reference Boeger, Vianna and Thatcher2006; Cohen et al., Reference Cohen, Justo and Kohn2013). Species from this class of Platyhelminthes are often used as a model system in studies that investigate host–parasite coevolution (Domingues and Boeger, Reference Domingues and Boeger2005; Mendlová and Šimková, Reference Mendlová and Šimková2014; Braga et al., Reference Braga, Razzolini and Boeger2015; da Graça et al., Reference da Graca, Fabrin, Gasques, Prioli, Balbuena, Prioli and Takemoto2018). Historical host–parasite associations between Monogenoidea and freshwater Neotropical fishes are fascinating due to the host's geographic isolation in hydrographic basins, which have complex biogeographical histories (Albert et al., Reference Albert, Tagliacollo and Dagosta2020), and the phylogenetic conservatism of host repertoires among the parasites (Braga et al., Reference Braga, Razzolini and Boeger2015).
Characithecium Mendoza-Franco, Reina & Torchin, 2009 (Dactylogyridae) is a genus of Monogenoidea with nine described species parasitizing the gills of freshwater fishes in Central (Mexico and Panama) and South America (Colombia, Brazil and Argentina). So far, species of Characithecium are known to occur on a few species of Astyanax [Astyanax aeneus (Günther, 1860), Astyanax ruberrimus Eigenmann, 1913 [synonym of Astyanax panamensis (Günther, 1864)], Astyanax lacustris (Lütken, 1875), Astyanax scabripinnis (Jenyns, 1842) and Astyanax bimaculatus (Linnaeus, 1758)], and on Psalidodon fasciatus (Cuvier, 1819) and Oligosarcus jenynsii (Günther, 1864) (Kritsky and Leiby, Reference Kritsky and Leiby1972; Gioia et al., Reference Gioia, Cordeiro and Artigas1988; Boeger and Vianna, Reference Boeger, Vianna and Thatcher2006; Rossin and Timi, Reference Rossin and Timi2015; Gallas et al., Reference Gallas, Calegaro-Marques and Amato2016).
Characithecium remained monotypic until Rossin and Timi (Reference Rossin and Timi2015) proposed a diagnostic amendment for the genus and a new species combination to accommodate Palombitrema chascomusensis Suriano, 1981 (=Characithecium chascomusensis). These authors also described four new species from the gills of O. jenynsii collected in the Chascomús lake and Nahuel Rucá lake, in the province of Buenos Aires (Argentina). Later, Gallas et al. (Reference Gallas, Calegaro-Marques and Amato2016) described Characithecium triprolatum Gallas, Calegaro-Marques & Amato, 2016, from the gills of Psalidodon aff. fasciatus and Astyanax jacuhiensis Cope, 1894 (=junior synonym of A. lacustris) from the Guaíba lake, in Rio Grande do Sul State (Brazil). Recently, new studies have described two new species of Characithecium occurring on the gills of Astyanax and Psalidodon species in Brazil. Da Silva et al. (Reference da Silva, da Silva and Yamada2021) described Characithecium bifurcuprolatum da Silva, da Silva & Yamada, 2021, and reported the occurrence of Characithecium costaricensis, both species found on the gills of A. bimaculatus in Northeast Brazil, and Zago et al. (Reference Zago, Franceschini, Abdallah, Müller, Azevedo and da Silva2021) described Characithecium paranapanemense Zago, Franceschini, Abdallahb, Müller & Azevedo, 2021 parasitizing the gills of Psalidodon paranae (Eigenmann, 1914) and Psalidodon bockmanni (Vari and Castro, 2007) collected in streams of the Middle Paranapanema river. This last article carried out morphological and molecular analyses to characterize the described species. In a molecular phylogeny using 28S rDNA, species of Dactylogyridae were included, as well as some samples of C. paranapanemense and C. costaricensis, where these two latter species were recovered as sister species in a clade closely related to species of Urocleidoides. However, a more comprehensive phylogeny for Characithecium, built to study the evolutionary history of the clade, is still needed.
The occurrence of Characithecium in phylogenetically close hosts led Rossin and Timi (Reference Rossin and Timi2015) to highlight the possibility of coevolution between the species of parasites and their hosts. Recent studies recovered (Astyanax (Andromakhe (Oligosarcus + Psalidodon))) composing the Gymnocharacini tribe within Stethaprioninae, a species-rich subfamily within Characidae (Mirande, Reference Mirande2020; Terán et al., Reference Terán, Benitez and Mirande2020). Oligosarcus is monophyletic, composed of 22 species, and distributed in river basins in southeastern South America (Ribeiro and Menezes, Reference Ribeiro and Menezes2015; Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019). On the contrary, Astyanax has long been recognized as a polyphyletic genus in Characidae, with some of its species having been recently transferred to the genera Psalidodon and Andromakhe to represent monophyletic genera (see Mirande, Reference Mirande2020; Terán et al., Reference Terán, Benitez and Mirande2020). Wendt et al. (Reference Wendt, Silva, Malabarba and Carvalho2019) recently studied the phylogenetic relationships, divergence times and biogeography of Oligosarcus species, including an extensive outgroup formed by several species currently referred to Astyanax and Psalidodon. According to this reconstruction, the most recent common ancestor of Oligosarcus inhabited drainages in the Brazilian crystalline shield (which included the Upper Paraná river basin and coastal region drainages of Brazil) in the Pliocene (~5 Ma), and the biogeographical history of this genus was influenced by sea-level changes during the Pleistocene and the formation of barriers (waterfalls) in the Paraná river basin (Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019).
The complex evolutionary history of Oligosarcus directly reflects the changes in drainage configurations that occurred in the southeastern region of South America (Ribeiro, Reference Ribeiro2006; Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019). Vicariant events were likely responsible for the current allopatric distribution of most lineages of Oligosarcus, and subsequent dispersion events resulted in secondary contact and sympatry in some groups of the genus (Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019). These events of separation and connection of lineages within Oligosarcus may have influenced the diversity and evolutionary history of the parasites occurring in these fishes. Furthermore, species of the clade containing Oligosarcus, Astyanax, Andromakhe and Psalidodon have relatively broad ecological requirements in freshwaters (Costa-Pereira et al., Reference Costa-Pereira, Araújo, Paiva and Tavares2016; Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019), with species variably occurring in distinct habitats (e.g. rivers, streams and lakes) and in distinct portions of drainages (e.g. headwaters and lowlands).
A wide range of evolutionary characteristics observed in these fishes and the complex host–parasite relationship make Characithecium and its host's excellent models for coevolutionary studies. Furthermore, a detailed study on the geographic distribution of Characithecium and the occurrence of these parasites in different hosts can be analysed following the assumptions of the Stockholm paradigm (Hoberg and Brooks, Reference Hoberg and Brooks2015). The Stockholm paradigm is a theoretical concept that uses several ecological processes, such as ecological fitting, oscillation and taxon pulse, to explain how the host–parasite relationship evolves, where new hosts (=resources) can be added or lost over evolutionary time. Thus, this new concept resolved the parasite paradox (Agosta et al., Reference Agosta, Janz and Brooks2010) since specialist parasites are widely found in nature even so, several phylogenetic and coevolutionary studies demonstrate high levels of host switching. After the proposal of the Stockholm paradigm, it was possible to understand better that parasites can promptly colonize new hosts when given an opportunity. Therefore, the broad ecologies and geographic distributions of the species of Oligosarcus, Astyanax, Andromakhe and Psalidodon may provide opportunities for parasites, including Characithecium, to colonize new hosts (Araujo et al., Reference Araujo, Braga, Brooks, Agosta, Hoberg, von Hartenthal and Boeger2015; Hoberg and Brooks, Reference Hoberg and Brooks2015; Brooks et al., Reference Brooks, Hoberg and Boeger2019; Agosta and Brooks, Reference Agosta and Brooks2020).
Hence, this study evaluates the evolutionary history and diversification of species of Characithecium. Phylogenetic relationships and species limits were evaluated based on molecular data, species distribution (both in geographical space and among hosts) and was tested if parasite prevalence could be predicted by ecological characteristics.
Materials and methods
Parasite sampling
The host sampling consisted mainly of numerous specimens from scientific collections which had already been collected previously. In addition, when necessary, new collections were carried out to cover sampling gaps. Therefore, the samples come from 2009 to 2019 and refer to different seasons of the year. Parasites collected from fish specimens were fixed and preserved in 96% alcohol for molecular analyses, and parasites found in specimens fixed in formalin 10% and preserved in 70% alcohol were used for morphological identification and to assemble permanent slides. Host specimens deposited in the following ichthyological collections were examined: Coleção de Peixes do Departamento de Zoologia e Botânica da Universidade Estadual Paulista, São José do Rio Preto (DZSJRP); Laboratório de Biologia e Genética de Peixes da Universidade Estadual Paulista, Botucatu (LBP); Museu de Ciências e Tecnologia da Pontifícia Universidade Católica do Rio Grande do Sul, Porto Alegre (MCP); Museu de Zoologia da Universidade Estadual de Londrina, Londrina (MZUEL); Coleção de Peixes do Departamento de Zoologia da Universidade Federal do Rio Grande do Sul, Porto Alegre (UFRGS) and Coleção Zoológica da Universidade Federal do Mato Grosso do Sul, Campo Grande (ZUFMS). Additional host specimens have recently been collected in field expeditions to fill gaps in host–species representation and their geographic distribution. These specimens were euthanized in a solution containing Eugenol (following Lucena et al., Reference Lucena, Calegari, Pereira and Dallegrave2013) and then fixed and preserved in 96% alcohol. Collection permits were given by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) to LRM (SISBIO No. 12038-1). Applicable institutional guidelines for the care and use of animals were followed and approved by the Ethics Committee of the Universidade Federal do Rio Grande do Sul (Porto Alegre, Brazil; CEUA-32283).
For evaluation of host–parasite relationships, we examined 351 specimens of fishes representing 17 species of Oligosarcus and 124 specimens representing nine species of Astyanax and six of Psalidodon from different geographic regions. Astyanax and Psalidodon belong to clades that are closely related and often sympatric to Oligosarcus species (Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019). For parasite removal, the gills were intensively washed with 96% alcohol bursts using a syringe, and whenever possible, gill arches were carefully dissected and analysed. Then, parasite specimens were stored in vials containing 96% alcohol and kept in a freezer at −4 °C. Part of these parasite specimens (fixed in formalin 10%) was mounted on permanent slides using Hoyer, and identified to the species level, using an Olympus BX51 microscope, following morphological characteristics given by Mendoza-Franco et al. (Reference Mendoza-Franco, Reina and Torchin2009), Rossin and Timi (Reference Rossin and Timi2015) and Gallas et al. (Reference Gallas, Calegaro-Marques and Amato2016). In addition to using the original descriptions, all the material was carefully compared with the voucher specimens of C. triprolatum (deposited in the helminthological collection of UFRGS) and from photos of the holotypes of the species C. chascomusensis, Characithecium chelatum, Characithecium longianchoratum, Characithecium robustum and Characithecium quadratum provided by Dr Lia Lunaschi, curator of the helminthological collection at Colecc de Invertebrados del Museo de La Plata, FCNyM-UNLP. Characithecium species were determined based on diagnostic characters such as the size and shape of the sclerotized parts of the attachment organ (haptor) and the reproductive organs (male copulatory complex and vaginal opening). Other parasite specimens (fixed in 96% alcohol) were used in molecular analysis, so they were mounted on a temporary glass slide containing glycerin and identified to the species level. Then, a permanent slide was designated as a paragenophore, to represent the sample used in molecular analyses. The paragenophores were mounted in Hoyer mounting medium (Humason, Reference Humason1979) and deposited in the helminthological collection of the Instituto Oswaldo Cruz, Rio de Janeiro, Brazil (CHIOC) and helminthological collection of the Instituto de Biociências da Universidade Federal do Rio Grande do Sul (UFRGS; Table S1).
DNA extraction, polymerase chain reaction (PCR) and sequencing
Most of the examined samples were from museum-preserved material, which allowed us to have extensive sampling, but required specific activities during molecular processing to avoid DNA loss. Characithecium specimens were removed and sequenced from Oligosarcus and Astyanax hosts given that the examined Psalidodon specimens were fixed only in formalin, making it difficult for DNA extraction. DNA was extracted from individual parasites (n = 35) according to the simplified method described by Tkach and Pawlowski (Reference Tkach and Pawlowski1999), which was designed to result in minimal DNA loss. The DNA of seven of the nine species described for Characithecium was extracted and amplified: C. costaricensis, C. chascomusensis, C. chelatum, C. longianchoratum, C. quadratum, C. robustum and C. triprolatum.
Furthermore, regarding the two species recently described for Characithecium, only C. paranapanemense has sequences of 28S rDNA deposited in GenBank and these sequences were properly included in the phylogenetic analysis (GenBank accession numbers: MZ408907, MZ408902 and MZ408908). Two ribosomal nuclear genes were amplified, 28S rDNA and 18S rDNA. The primers C1 (5′ ACCCGCTGAATT TAAGCAT 3′) and C3 (5′ CTCTTCAGAGTACTTTTCAAC 3′) were used to amplify a fragment of approximately 400 bp of 28S rDNA (Mollaret et al., Reference Mollaret, Lim and Justine2000). After several efforts to amplify the 18S rDNA using primers available in the literature (e.g. Littlewood and Olson, Reference Littlewood, Olson, Littlewood and Bray2001), we were not successful. Therefore, a new primer was designed to amplify the 18S rDNA region. These new primers – 18S rDNA: 188F (5′ TGACGTTGGATGTCAGACGG 3′) and 18S rDNA: 486R (5′ TAGTTTGTC TGGCGACGGTC 3′) – were used to amplify a fragment of approximately 478 bp of 18S rDNA. These primers were developed based on a sequence of the Dactylogyridae Diaphorocleidus armillatus Jogunoori, Kritsky, and Venkatanarasaiah, 2004 (GenBank accession number: KT597997.1). Primer3Plus software (Untergasser et al., Reference Untergasser, Nijveen, Rao, Bisseling, Geurts and Leunissen2007) was used, and the quality of the primer was tested in software NetPrimer (http://www.premierbiosoft.com/netprimer/netprlaunch/netprlaunch.html).
The PCR programme for 28S rDNA was as follows: 5 min at 95 °C, followed by 40 cycles of 1 min at 94 °C, 1 min at 45 °C, 2 min at 72 °C and completed with 7 min at 72 °C. The PCR programme for 18S rDNA was as follows: 5 min at 95 °C, followed by 40 cycles of 1 min at 95 °C, 45 s at 50 °C, 1 min at 72 °C and finally 5 min at 72 °C. Each amplification reaction contained 3–5 μL template DNA, 3 mm MgCl2, 1× PCR buffer (Invitrogen, Massachusetts, USA), 0.5 pmol each primer, 0.4 mm dNTP and 1 U platinum Taq polymerase (Invitrogen) in a total volume of 25 μL. PCR products were checked by electrophoresis in agarose gel, purified using ExoSap (Exonuclease I and Shrimp Alkaline Phosphatase GE Healthcare®, Piscataway, NJ, USA) and sequenced in both directions by ACTGene (Porto Alegre, Brazil). Forward and reverse sequences were visually inspected, edited using software Geneious 8.0 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Mentjies and Drummond2012) and aligned using default parameters of the algorithm MAFFT (Katoh et al., Reference Katoh, Misawa, Kuma and Miyata2002), available on the GUIDANCE server (http://guidance.tau.ac.il/; Penn et al., Reference Penn, Privman, Ashkenazy, Landan, Graur and Pupko2010), that guides the removal of parts of the alignment that showed low reliability. The sequences of 28S rDNA and 18S rDNA of Characithecium species were deposited in GenBank (Table S1). The pairwise genetic distances were calculated in Geneious 8.0 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Mentjies and Drummond2012) using the Tamura–Nei model (Tamura and Nei, Reference Tamura and Nei1993; Table S2).
Phylogenetic reconstruction and divergence time estimation
Bayesian inference was performed using BEAST2 v2.5.1 (Bouckaert et al., Reference Bouckaert, Heled, Kühnert, Vaughan, Wu, Xie, Suchard, Rambaut and Drummond2014) to estimate phylogenetic relationships of the gene tree for 28S rDNA (the most densely sampled marker). Nucleotide substitution models for 28S rDNA and 18S rDNA genes were evaluated using PartitionFinder v1.1.1 (Lanfear et al., Reference Lanfear, Calcott, Ho and Guindon2012). For the gene tree analysis of the 28S rDNA, the birth–death model was set as a tree prior, and the relaxed lognormal clock was used as the clock model. The analysis ran for 10 000 000 generations, sampling every 1000 generations and species of Dactylogyridae selected from GenBank were used as outgroup (Table S1).
Using this single gene (28S rDNA) dataset, a molecular time divergence analysis was conducted in BEAST v2.5.1 (Bouckaert et al., Reference Bouckaert, Heled, Kühnert, Vaughan, Wu, Xie, Suchard, Rambaut and Drummond2014). For that, we used the evolutionary rate of the 28S rDNA proposed to Proseriata (Platyhelminthes; Scarpa et al., Reference Scarpa, Cossu, Sanna, Lai, Norenburg, Curini-Galletti and Casu2015). A relaxed lognormal clock model was set, with an evolutionary rate of 0.005 mutations per million years for the 28S rDNA. The birth–death model was used as a tree prior (Nee et al., Reference Nee, May and Harvey1994).
Species Tree analysis using both markers (28S rDNA and 18S rDNA) was conducted. Specimens of Jainus and Cacatuocotyle were included as outgroups. The birth–death model was set as the tree prior, and the relaxed clock was configured as the clock model. The analysis was conducted with two runs of four chains conducted simultaneously for 5 000 000 generations with a sample frequency of 500 generations. The StarBEAST 2.5 template (Heled and Drummond, Reference Heled and Drummond2010) was used and the relaxed clock and birth–death tree models were linked to 28S rDNA and 18S rDNA datasets. The Species Tree analysis was conducted with two different approaches. First, an analysis without prior calibrations for date estimates on internal nodes. Then, an analysis using prior calibration dates on nodes based on divergence time estimations from the 28S rDNA dataset (see divergence time estimate analysis above). For that, three nodes that were congruent with the gene tree analysis (28S rDNA) were dated using minimum age with standard deviation and using normal priors in order to calibrate the respective nodes as follows: (1) Characithecium node (12.20 ± 2.6 Ma of standard deviation), (2) the node formed by C. triprolatum, C. quadratum and C. paranapanemense (1.46 ± 0.6 Ma standard deviation) and (3) the node formed by C. costaricensis (C. longianchoratum (C. chelatum (C. chascomusensis + C. robustum))) (6.08 ± 1.8 Ma standard deviation). The minimum age and standard deviation were obtained from the 28S rDNA tree dated from the evolutionary rate of this marker.
Morphological diagnostic traits of each species (Mendoza-Franco et al., Reference Mendoza-Franco, Reina and Torchin2009; Rossin and Timi, Reference Rossin and Timi2015; Gallas et al., Reference Gallas, Calegaro-Marques and Amato2016) were the criteria for grouping specimens into putative species in the Species Tree analysis. The analysis ran for 15 000 000 generations with a sample frequency of 1500 generations. All these analyses were implemented with XSEDE (3.2.6) in the CIPRES portal (Miller et al., Reference Miller, Pfeiffer and Schwartz2010).
For all the Bayesian analyses mentioned above, we inspected stationary posterior probabilities using Tracer v1.6 (Rambaut et al., Reference Rambaut, Suchard, Xie and Drummond2014) and checked for effective sample size's above 200. The first 10% of the trees were discarded as burn-in, and the remaining trees were summarized using the maximum clade credibility tree function in TreeAnnotator 2.4.3 (Bouckaert et al., Reference Bouckaert, Heled, Kühnert, Vaughan, Wu, Xie, Suchard, Rambaut and Drummond2014).
Species delimitation
Species-delimitation analyses were carried out and contrasted with the current proposed morphological delimitations, using variable characteristics within Characithecium, which distinguish one species from another (summarized in Table S3). We observed the morphology (shape of the sclerotized pieces) from the collected specimens (our examined material) and compared them with the literature information (Mendoza-Franco et al., Reference Mendoza-Franco, Reina and Torchin2009; Rossin and Timi, Reference Rossin and Timi2015; Gallas et al., Reference Gallas, Calegaro-Marques and Amato2016). Species-delimitation analyses using a single locus were carried out with the generalized mixed-yule coalescent (GMYC) method (Pons et al., Reference Pons, Barraclough, Gomez-Zurita, Cardoso, Duran, Hazell, Kamoun, Sumlin and Vogler2006; Fujisawa and Barraclough, Reference Fujisawa and Barraclough2013) and the Bayesian implementation of the Poisson tree processes (bPTP) method (Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013). According to Zhang et al. (Reference Zhang, Kapli, Pavlidis and Stamatakis2013), these two methods of species delimitation differ in that GMYC uses branch lengths (timed divergences) to identify when divergence times more closely resemble coalescence events rather than speciation events, while bPTP uses the number of substitutions. As the input tree for the GMYC, we used the summarized ultrametric tree that was reconstructed using the 28S rDNA gene in BEAST2 v.2.5.1. The GMYC analysis was carried out in the R package ‘Splits’ (Ezard et al., Reference Ezard, Fujisawa and Barraclough2009) with a single threshold. The bPTP analysis was carried out on the online server (https://species.h-its.org/) using the unrooted tree, following the default parameters (with 100 000 generations), and using the summarized (not ultrametric) 28S rDNA tree reconstructed using MrBayes 3.2.2 (Ronquist et al., Reference Ronquist, Teslenko, Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012). For this tree, we set K80 + G as the nucleotide substitution model (as proposed by PartitionFinder) and conducted two simultaneous runs of four chains over 10 000 000 generations, sampling every 1000 generations.
Occurrence and ecological traits of Characithecium
After the collection and subsequent species determination of parasites, we characterized each species of Characithecium based on: (1) host genus; (2) host species; (3) prevalence in each host species, i.e. the percentage of examined host specimens that contained the focal parasite species (Bush et al., Reference Bush, Lafferty, Lotz and Shostak1997); (4) parasite geographic distribution, which includes country, state, river basin, freshwater ecoregion and if it belongs to coastal and/or continental basins; (5) altitude of occurrence (in metres) and (6) categorical habitat type (river, stream, lake or a combination of those).
It was tested whether some biotic and abiotic variables explain the prevalence of each species of Characithecium. Generalized linear models (GLMs) were used for this analysis, using the binomial distribution (recommended for data with proportions, such as prevalence). In total, 13 models were created (M1–M13) with interactions between one or more of the following four variables: (1) geographic distribution – ecoregion, (2) habitat type, (3) altitude class and (4) host genus – as a proxy for host phylogeny (Table 1; Fig. 1). For the GLM analysis, the altitude values were divided into four classes that were discretized based on gaps in the observed altitude values among hosts and parasites, generating the following altitude classes: class 1 = 0–100 m, class 2 = 101–400 m, class 3 = 401–800 m, class 4 = more than 801 m. We followed the Freshwater Ecoregions of the World (FEOW) proposed by Abell et al. (Reference Abell, Thieme, Revenga, Bryer, Kottelat, Bogutskaya, Coad, Mandrak, Balderas, Bussing, Stiassny, Skelton, Allen, Unmack, Naseka, Ng, Sindorf, Robertson, Armijo, Higgins, Heibel, Wikramanayake, Olson, López, Reis, Lundberg, Sabaj and Petry2008) for ecoregion delimitation. In addition, we tested the null model (M0), where the prevalence of the parasite species was not associated with any of the above variables. We used Akaike's information criterion (AICc) to select the model(s) that best explained the observed patterns, where models with ΔAICc ⩽ 2 were considered to perform equally well (Burnham and Anderson, Reference Burnham and Anderson2002). Finally, we applied analysis of variance (ANOVA) with error type III (ideal for unbalanced data) to test the significance for each best model selected, using an α-value of 0.05.

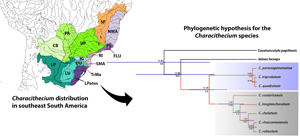
Fig. 1. Schematic representation of variables used to explain parasite prevalence from GLM analysis, with competing models explaining the parasite species prevalence. Map locating the ecoregions, which correspond to the study area in southeastern South America. Ecoregion abbreviations: LPatos, Laguna dos Patos; TrMa, Tramandaí-Mampituba; SMA, southeastern Mata Atlântica; RI, Ribeira de Iguape; FLU, Fluminense; PS, Paraíba do Sul; NMA, northeastern Mata Atlântica; SF, São Francisco; UP, Upper Paraná; LP, Lower Paraná; IG, Iguaçu; PA, Paraguay; CB, Chaco; UU, Upper Uruguay; LU, Lower Uruguay.
Table 1. Models created with ecological variables used to explain parasite prevalence from GLM analysis

Results
Phylogenetic relationships, divergence time estimates and species delimitations
The fragment of the 28S rDNA (~400 bp) was composed of 38 individuals of Characithecium, while for 18S rDNA, which corresponded to the longest region (~478 bp), was successfully amplified for only nine individuals, representing four species of Characithecium. The K80 + G model was selected as the best substitution model for the 28S rDNA, while the TrNef model was selected for the 18S rDNA. The 28S rDNA fragment presented greater genetic variation compared to 18S rDNA, with 141 and 25 mutations and 92 parsimony informative sites contrasting with only 19 in the 18S rDNA. The mean genetic distance (P-distance) between the sequences of 28S rDNA was 0.101 contrasting with 0.024 in the 18S rDNA and base frequency was A = 27.9%, C = 20.3%, G = 28.6%, T = 23.2% and for 28S rDNA A = 22.9%, C = 20.4%, G = 29.0% and T = 27.7%. Overall, intraspecific uncorrected P-distances ranged from 0 to 1.15%, while the interspecific distances ranged from 0.31 to 14.12% (Table S2).
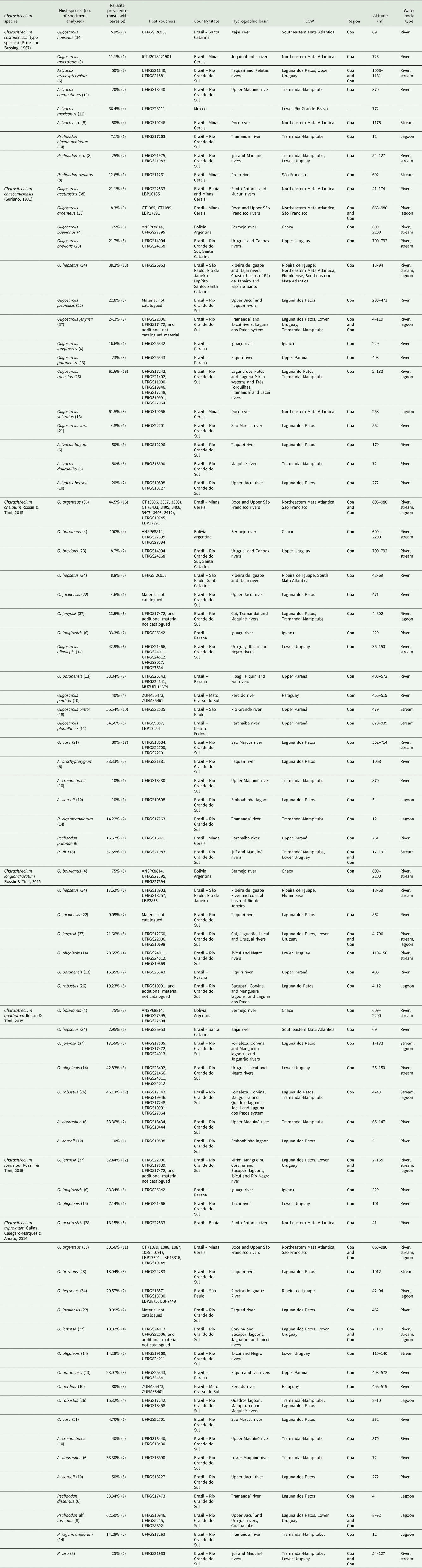
The phylogenetic reconstruction based on the 28S rDNA recovered Characithecium as monophyletic with high node support (PP = 0.92; Fig. 2). Given the available data, Characithecium was estimated to be a sister group to Jainus hexops, and these two genera are a sister group to a large clade of Dactylogyridae (Fig. 2). Within Characithecium, specimens were grouped in reciprocally monophyletic groups that supported the morphologically determined species relationships (Fig. 3), except for three species (C. triprolatum, C. quadratum and C. paranapanemense). Individuals from these three species share a most recent common ancestor (with high support), but samples show no reciprocal monophyly. We recovered a larger clade with the remaining five species of the genus with high support (PP = 1.0), being composed of C. costaricensis + (C. longianchoratum + (C. chelatum + (C. robustum + C. chascomusensis))). The Species Tree analysis revealed a similar topology of the gene tree analysis (28S rDNA), with high support values for most clades. The Species Tree analysis revealed C. paranapanemense as a sister species of the clade composed of C. triprolatum and C. quadratum (PP = 1.0; Fig. 4).

Fig. 2. Calibrated ultrametric hypothesis for species of Characithecium, together with other Dactylogyridae species, based on Bayesian inference to 28S rDNA fragment. Time estimation (median age in Ma) is represented by values above branches and posterior probabilities above 0.5 are shown below branches.

Fig. 3. Phylogenetic hypothesis for Characithecium species based on 28S rDNA (left phylogram). Species delimitations on the right using the GMYC model and the bPTP model. Terminals in the Characithecium phylogeny include the name of the species of Characithecium and the name of the host in which the parasite individual was found. Posterior probabilities above 0.5 are shown below branches.

Fig. 4. Time-calibrated Species Tree hypothesis for Characithecium species based on 28S rDNA and 18S rDNA. Values below branches represent posterior probabilities. Time estimation (median age in Ma) is represented by values above branches. Blue bars represent the variation (95%) of the estimated dates.
The non-monophyly of C. paranapanemense, C. triprolatum and C. quadratum, observed in the molecular analysis, is inconsistent with the morphological data (Table S3). Characithecium triprolatum and C. quadratum differ in several ways, such as posteromedial projection in ventral bar (present in C. triprolatum vs absent in C. quadratum); medial suture in ventral bar (absent in C. triprolatum vs present in C. quadratum); accessory piece of male copulatory organ (MCO) shape (pincer-shaped in C. triprolatum vs clamp-shaped in C. quadratum) and vaginal opening (ventral in C. triprolatum vs marginal in C. quadratum; Table S3). On the contrary, C. paranapanemense have a similar morphology to C. triprolatum, but differs from this species mainly because it has a ventral bar with irregular anterior margin and a large posteromedial projection.
The divergence time estimates recovered that the origin of Characithecium diversification (i.e. its first-cladogenetic event) is dated back to approximately 11 Ma (95% highest posterior density (HPD) = 17.03–7.63 Ma; Figs 2 and 4), which corresponds to the middle Miocene. It was estimated that the clade C. paranapanemense (C. triprolatum + C. quadratum) evolved approximately 0.96 Ma (95% HPD = 1.95–0.13 Ma). Characithecium costaricensis diverged from its sister group at around 5.58 Ma (95% HPD = 8.99–2.95 Ma), and the divergence between C. robustum and C. chascomusensis was estimated at approximately 0.66 Ma (95% HPD = 1.95–0.01 Ma).
The results of the species-delimitation methods (GMYC and bPTP) based on a single locus did not recover the eight morphological entities presently recognized (Fig. 3). The species delimitation using the gene 28S rDNA supports four species in the GMYC analysis, while the bPTP analysis revealed six species (Fig. 3). Both methods indicate that C. paranapanemense, C. triprolatum and C. quadratum may represent the same taxonomical unit. In addition, the GMYC, contrasting with the bPTP, did not recover C. chelatum, C. robustum and C. chascomusensis as distinct species (Fig. 3).
Characithecium occurrences and ecological traits
New hosts and expanded geographic distributions were detected for the species of Characithecium (Table 2). This extensive host repertoire for the parasite genus contains 32 fish species, with most interactions occurring at low prevalence rates. Two species of parasites occur exclusively in the members of Oligosarcus (C. longianchoratum and C. robustum; Table 2). In general, Characithecium species have higher prevalence rates in the species of Oligosarcus compared to that in Astyanax or Psalidodon species, and each parasite species usually has a high prevalence in a single host species among all observed host species (Table 2).
Table 2. Species of Characithecium parasitizing gills of Oligosarcus, Astyanax and Psalidodon species, with prevalence, ecology and geographic distribution of hosts in South and Central America, and host voucher where the parasite was found. m, metres; Coa, coastal basins; Con, continental basin according to Wendt et al. (Reference Wendt, Silva, Malabarba and Carvalho2019); FEOW, Freshwater Ecoregions of the World according to Abell et al. (Reference Abell, Thieme, Revenga, Bryer, Kottelat, Bogutskaya, Coad, Mandrak, Balderas, Bussing, Stiassny, Skelton, Allen, Unmack, Naseka, Ng, Sindorf, Robertson, Armijo, Higgins, Heibel, Wikramanayake, Olson, López, Reis, Lundberg, Sabaj and Petry2008)
GLM analyses recovered that prevalence can be best explained by biotic and abiotic variables (Table 3). Host genus shown to be the variable that best explains the distribution of C. costaricensis, with the highest prevalence being found in Astyanax species (M3 and M4; Tables 2 and 3). The four variables together (M1 = host genus, altitude class, habitat type and ecoregion) were shown to better explain the distribution of C. chascomusensis (65%), with the highest prevalence being found in Oligosarcus species at higher altitudes (i.e. class 3 = more than 401 m), and in rivers in the coastal region of Brazil. Similarly, M1 was the model that best explained the distribution of C. quadratum (100%), with a higher prevalence in Oligosarcus, with distribution in La Plata river drainages.
Table 3. Best models selected by the GLM analysis that explain Characithecium prevalence

AICc, Akaike information criterion; ΔAICc, delta AICc; weight, weight of each model in the analysis.
Chi-squared is the statistic test used in ANOVA.
Significant values: ***P ⩽ 0.001; *P ⩽ 0.05.
The prevalences of C. robustum (76%) and C. triprolatum (73%) were best explained by habitat type and ecoregion (M10; Table 3). The high prevalence of C. robustum was associated with rivers in the Iguaçu ecoregion, whereas C. triprolatum presented higher prevalences in rivers in the Paraguay ecoregion. The high prevalence for C. longianchoratum was best explained by M6, associated with the Oligosarcus species and distributed in La Plata river drainages.
In addition, the GLM analysis revealed that altitude class and ecoregion better explained the distribution of C. chelatum (M3 = 63%), with higher prevalence at high altitudes (i.e. more than 401 m) and in La Plata river and Laguna dos Patos drainages (Tables 2 and 3).
Discussion
Molecular phylogeny and species delimitations
The phylogenetic structure reconstructed for the available sequences of Characithecium spp. recognized a major clade of parasite species, which was composed of five of the eight Characithecium species studied here, which agree with the previous morphological delimitations (Rossin and Timi, Reference Rossin and Timi2015; Gallas et al., Reference Gallas, Calegaro-Marques and Amato2016; Zago et al., Reference Zago, Franceschini, Abdallah, Müller, Azevedo and da Silva2021).
The species delimitations based on GMYC and bPTP support fewer species compared to the morphological delimitation based on a combination of diagnostic characters. This result may be a consequence of the low genetic variability of ribosomal genes for the level of resolution necessary to discriminate closely related entities (e.g. when compared to mitochondrial genes; Ruttkay et al., Reference Ruttkay, Solignac and Sperlich1992; Lemey et al., Reference Lemey, Salemi and Vandamme2009). A larger number of species was estimated by bPTP compared to GMYC, which can be explained by the difference in how these species-delimitation methods work. While bPTP uses the number of nucleotide substitutions to model species boundaries, GMYC uses information from the distribution of divergence times, which can be influenced by how the phylogeny was time-calibrated (Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013). A recent study found contrasting results for nematode parasites, where both models (GMYC and bPTP) recovered a larger number of species than morphological delimitation (Qing et al., Reference Qing, Bert, Gamliel, Bucki, Duvrinin, Alon and Miyara2019). This later study used, in addition to 18S rDNA and 28S rDNA, fragments of internal transcribed spacer (ITS) and cytochrome C oxidase subunit 1 (COI), which may have contributed to recognizing putative genetic variability (Qing et al., Reference Qing, Bert, Gamliel, Bucki, Duvrinin, Alon and Miyara2019). ITS and COI are known to have greater variability than ribosomal fragments (Bueno-Silva, Reference Bueno-Silva2012; Vanhove et al., Reference Vanhove, Tessens, Schoelinck, Jondelius, Littlewood, Artois and Huyse2013), where many studies also observed divergence between morphological and molecular data. For Monogenoidea, the vast majority of these studies reports a cryptic diversity, which is accessed after using more variable genes (Benovics et al., Reference Benovics, Desdevises, Šanda, Vukić, Scheifler, Doadrio, Sousa-Santos and Šimková2020; Ondračková et al., Reference Ondračková, Seifertová, Bryjová, Leis and Jurajda2020; Pinacho-Pinacho et al., Reference Pinacho-Pinacho, Calixto-Rojas, García-Vásquez, Guzmán-Valdivieso, Barrios-Gutiérrez and Rubio-Godoy2021).
The recent diversification of some groups of Characithecium and the divergence between C. paranapanemense, C. triprolatum and C. quadratum are estimated to have occurred during the Pleistocene, which may contribute to these species going unrecognized by the delimitation methods using molecular data. Despite putative recent speciation, several morphological characters can distinguish C. triprolatum and C. quadratum, such as vaginal opening position (e.g. ventral in C. triprolatum vs marginal in C. quadratum) and characters related to the ventral bar morphology (e.g. posteromedial projection and suture in the ventral bar and ventral bar shape; Rossin and Timi, Reference Rossin and Timi2015; Gallas et al., Reference Gallas, Calegaro-Marques and Amato2016). On the contrary, C. paranapanemense has a similar morphology to C. triprolatum. However, according to Zago et al. (Reference Zago, Franceschini, Abdallah, Müller, Azevedo and da Silva2021), these species differ mainly concerning the MCO (with base possibly fused to the proximal portion of the dorsal subunit of the accessory piece in C. triprolatum) and the shape of the ventral bar, whereas C. paranapanemense has a ventral bar with an irregular anterior margin and a large posteromedial projection while C. triprolatum has a ventral bar with a regular anterior margin and a short median projection. Thus, we do not propose any nomenclatural change (e.g. synonymization of C. paranapanemense, C. triprolatum and C. quadratum) because morphological and molecular data may reach congruence when a wider variety of genetic markers are used in the study of Characithecium species. Furthermore, future analyses including C. bifurcuprolatum may provide more information on genetic data and present a new hypothesis of relationship.
Ecological factors influencing Characithecium prevalence
Characithecium spp. parasitize species of Oligosarcus, Astyanax and Psalidodon, and depict a wide geographical distribution throughout drainages in southeastern South America. We report the new occurrence of Characithecium in 28 additional host species. Characithecium seems to explore similar resources since it occurs in phylogenetically close hosts (Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019; Mirande, Reference Mirande2020; Terán et al., Reference Terán, Benitez and Mirande2020).
Until recently, Characithecium has been observed only in a few species of Astyanax and only one species of Oligosarcus and Psalidodon. Species of this genus have never been reported in the species of other host fish genera within Characidae, nor of other fish families or orders (Boeger and Vianna, Reference Boeger, Vianna and Thatcher2006; Cohen et al., Reference Cohen, Justo and Kohn2013). This previous knowledge on the distribution of Characithecium species suggested a high host specificity and strong evidence of host–parasite coevolution (see Rossin and Timi, Reference Rossin and Timi2015). However, the greater sampling effort of the present study revealed that all species of Characithecium can use a wide range of host species, sharing members of the closely related species of Astyanax, Psalidodon and Oligosarcus. These host–parasite associations seem to be consistent with the results found by Mendoza-Palmero et al. (Reference Mendoza-Palmero, Rossin, Irigoitia and Scholz2020), which studied a new species of Ameloblastella and other dactylogyrids species parasitizing Neotropical catfish (e.g. species of Pimelodidae) and recovered that the parasites do not exhibit strict host–parasite associations at the genus or species level, but rather at the family or subfamily level.
We also found contrasting levels of prevalence values within Characithecium spp. on distinct host species. High prevalence is generally explained with distinct ecological and geographical variables, such as host genus, habitat type and ecoregions. Characithecium spp. tended to be more prevalent when associated with Oligosarcus species and with certain ecological conditions of their hosts (such as inhabiting rivers or high altitudes), but also with other variables considered (e.g. ecoregion distribution). These results reveal the ability of these parasites to explore a diverse environment that is variable in biotic (e.g. hosts) and abiotic features. The absence of a clear pattern between the prevalence and ecological and geographical aspects suggests that species of Characithecium are ecologically flexible.
Several studies have demonstrated the great flexibility of parasites in exploring a wide range of resources and with different ecological predictors (Braga et al., Reference Braga, Razzolini and Boeger2015; Mendoza-Palmero et al., Reference Mendoza-Palmero, Rossin, Irigoitia and Scholz2020). Many studies have already investigated the factors that could be related to different abundances in monogenoid species, with body size and geographic distance from hosts being correlated with variations in abundance (Poulin and Justine, Reference Poulin and Justine2008; Poulin et al., Reference Poulin, Blanar, Thieltges and Marcogliese2011a). In the same way, a recent study sought to understand the variation of ecological parameters (e.g. abundance and prevalence of parasite) of Aglaiogyrodactylus spp. (Gyrodactylidae) among host species (fishes of Loricariidae family; Patella et al., Reference Patella, Brooks and Boeger2017). According to the authors, differences in host use parameters are directly related to the level of compatibility, that is, the capacity of the parasite species to establish its population in each host. Therefore, more compatible host species are more intensively parasitized, and alternative hosts presented abundances at least twice as small as the first ones (Patella et al., Reference Patella, Brooks and Boeger2017). Lower abundances apparently indicate less compatible host species (Patella et al., Reference Patella, Brooks and Boeger2017).
Although host specificity has been defined historically as the number of hosts used by the parasite (Poulin, Reference Poulin1992; Kearn, Reference Kearn1994; Huyse and Volckaert, Reference Huyse and Volckaert2005; Poulin et al., Reference Poulin, Krasnov and Mouillot2011b), it is now recognized by how parasites are specialized to the resource(s) their hosts represent and not only to their taxonomy or phylogeny (although these can represent adequate proxies to the quality and quantity of resources) (Brooks and McLennan, Reference Brooks and McLennan2002; Brooks et al., Reference Brooks, Hoberg and Boeger2019; Agosta and Brooks, Reference Agosta and Brooks2020). Most elements that compose these resources are likely heritable and conserved in closely related hosts (see also Brooks and Agosta, Reference Brooks and Agosta2012). Hence, the present study indicates that species of Characithecium exploit resources that are unique to the host clade they parasitize (Astyanax (Andromakhe (Psalidodon + Oligosarcus))) while occurring under a broad range of environmental conditions. A comprehensive analysis of host–parasite networks in a geographic context should provide a complete understanding of the historical processes associated with the evolution of this host–parasite system.
The discussion above is directly linked to the Stockholm paradigm, representing an alternative theoretical framework for understanding host–parasite interactions (Brooks et al., Reference Brooks, Hoberg, Boeger, Gardner, Galbreath, Herczeg, Mejía-Madrid, Rácz and Dursahinhan2014, Reference Brooks, Hoberg and Boeger2019). This paradigm purports that parasites are resource trackers (as opposed to trackers of the host phylogeny) and diversify mostly by host switching. The Stockholm paradigm suggests that a parasite can readily infect compatible hosts by ecological fitting (Janzen, Reference Janzen1985) if the encounter between the symbionts presents itself (see also Araujo et al., Reference Araujo, Braga, Brooks, Agosta, Hoberg, von Hartenthal and Boeger2015). Compatibility (or capacity) and opportunity determine the realized fitness space (Brooks et al., Reference Brooks, Hoberg and Boeger2019; =the operative environment of Agosta and Klemens, Reference Agosta and Klemens2008) of consumer species, which is envisioned as the host-repertoire of a parasite species. Hence, colonization of new host species may result from factors such as phylogeny (a proxy to compatibility) and geographic proximity, as well as biological and ecological conditions (the later representing determinants of opportunity).
Indeed, monogenoids (Dactylogyridae) that parasitize Neotropical freshwater fishes show different association patterns depending on the host taxonomic group (Braga et al., Reference Braga, Razzolini and Boeger2015). For instance, within the most species-rich host group, the Characiformes, parasites generally share hosts that are closely related phylogenetically (i.e. likely as a proxy to similar resources). In contrast, for another group, the Siluriformes, environmental characteristics and geographical proximity (i.e. determinants of opportunity) presented a more significant effect on the level of host sharing (Braga et al., Reference Braga, Razzolini and Boeger2015). Therefore, the association between Characithecium species and their hosts seems to follow the pattern observed for other monogenoids in Characiformes, in which a close phylogenetic relationship of their hosts has an important effect.
The wide geographical distribution of Characithecium species may be associated with the evolutionary history of the hosts and dispersal events that may, in turn, have provided various opportunities for new host colonization. The host Oligosarcus, for example, is estimated to have an ancestral area in the Brazilian crystalline shield, and from there, two clades diverged and dispersed across La Plata river system tributaries and coastal drainages of southeastern South America (Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019). GLM analyses indicate that the high prevalence rates were generally associated with ecoregions that compose tributaries of the La Plata river basin (e.g. Iguaçu, Paraguay and Chaco), being the region marked by the diversification of one clade of Oligosarcus at approximately 3 Ma (Wendt et al., Reference Wendt, Silva, Malabarba and Carvalho2019). Subsequent dispersal events of Oligosarcus spp. from continental basins to coastal basins and from coastal basins to continental basins, dated to late Pleistocene, may have created new opportunities for the parasites to colonize new hosts on new geographic areas, even if the hosts currently have low prevalence rates.
Finally, the diversification of Characithecium and its relationship with its hosts demonstrated numerous particularities, which are added to the complex formation of drainages in South America. Such levels of complexities demonstrate the need to carry out increasingly interactive studies, including different types of information (morphological, molecular, ecological and others), and use increasingly complete databases to understand the evolutionary history of species and their associations. Then, the present study used a wide range of samples from many scientific collections that provided the largest number of host–parasite associations from many locations. Therefore, biological collections were very important to reconstruct a comprehensive phylogenetic relationship for Characithecium and to describe a broad host–parasite relationship in rivers of South America.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022000221.
Acknowledgements
We acknowledge all the financial institutions (listed below) that contributed to this study being carried out. We thank Dr Lia Lunaschi, the curator of the helminthological collection at Colección de Invertebrados del Museo de La Plata, for providing photos of the holotypes of the several Characithecium species. We thank Dr Marcelo Knoff, the curator of the helminthological collection of the Instituto Oswaldo Cruz, Rio de Janeiro, Brazil (CHIOC), for providing Characithecium vouchers in this important zoological collection. We thank the curators and personnel of the following ichthyological collections for providing material: F. Langeani (DZSJRP); C. Oliveira (LBP); C. Lucena (MCP); F. Jerep and O. Shibatta (MZUEL) and F. Carvalho (ZUFMS). We also thank V. Tkach, J. Weckstein and L. Bachmann for help and comments provided on the molecular work on parasites, host–parasite relationships and for many other discussions. We thank A. Ferrari, B. Sidlaukas and R. Vianna for comments on an early version of this manuscript.
Author contributions
E. W. W., L. R. M. and T. P. C. conceived and designed the study. E. W. W. and T. P. C. conducted data gathering. L. R. M. and T. P. C. checked the fish identifications and taxonomy. E. W. W. and W. A. B. checked the parasite identifications and taxonomy. E. W. W., M. P. B. and W. A. B. designed analyses of host–parasite association. E. W. W., M. P. B., M. L. and T. P. C. conducted statistical analyses. E. W. W., L. R. M., M. P. B., W. A. B., M. L. and T. P. C. wrote the article.
Financial support
This study was funded by CAPES with a doctorate scholarship and was partially funded by Systematics Research Fund 2017/18, both granted to E. W. W. Field trips for collecting hosts were partially funded by a CNPq Universal grant (401204/2016-2) to L. R. M. T. P. C., during most of the duration of this project, was funded by CAPES with a PNPD postdoctoral fellowship.
Conflict of interest
None of the authors have any conflicts of interest.
Ethical standards
This project was approved by the institution's ethics committee (No. 32283) and followed all the proposed recommendations.