Introduction
Climate change may lead to the increased transmission of, and shift the spatial distribution of infectious diseases (Patz et al., Reference Patz, Epstein, Burke and Balbus1996). In combination with other anthropogenic factors such as increased housing, settlements, and environmental pollution, extensive tropical urbanization, colonization, and changing lifestyle patterns, these changes have produced new breeding areas for vectors of many diseases, with significant implications for human and animal health (Weaver and Reisen, Reference Weaver and Reisen2010; Hongoh et al., Reference Hongoh, Berrang-Ford, Scott and Lindsay2012; Carvalho et al., Reference Carvalho, Rangel and Vale2017). Mosquitoes are a major public health concern, as they play a vital role in transmitting human and animal diseases (Tandina et al., Reference Tandina, Doumbo, Yaro, Traoré, Parola and Robert2018). Culex pipiens L. (Culicidae: Diptera) is a cosmopolitan mosquito causing nuisance and transmitting several significant pathogens such as West Nile virus, St. Louis encephalitis virus, Zika virus, Sindbis/Sindbis-like viruses, Usutu flavivirus, avian malaria (Plasmodium spp.) and filarial worms (Farajollahi et al., Reference Farajollahi, Fonseca, Kramer and Marm Kilpatrick2011).
Research on avian malarial parasites has been heavily focused towards detection and understanding avian host–parasite relationships. In contrast, few studies have been carried out on vector–parasite relationships, due to lack of available colonies of many vector mosquito species and because cultures of avian Plasmodium species and lineages are not easily available (Valkiūnas, Reference Valkiūnas2011). The investigation of vector–parasite relationship requires a source of infective avian blood, which is not available for many of the >55 morphospecies of avian plasmodium species described to date (Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018).
As is the case for most of the African continent (Valkiūnas, Reference Valkiūnas2005), few studies have examined the blood parasites of birds in Egypt. More than 150 species of migratory birds visit Egypt annually, in addition to 350 resident species of birds (Ibrahim, Reference Ibrahim2011). Despite this, most studies on avian malarial parasites in Egypt were conducted more than five decades ago (Haiba, Reference Haiba1948; Mohammed, Reference Mohammed1958a, Reference Mohammedb; Guindy et al., Reference Guindy, Hoogstraal and Mohammed1965). Only one study that detected the Plasmodium lineages PADOM02, PADOM16 and SGS1 in sparrows (Passer domesticus) sampled at Luxor (Marzal et al., Reference Marzal, Ricklefs, Valkiūnas, Albayrak, Arriero, Bonneaud, Czirják, Ewen, Hellgren, Hořáková, Iezhova, Jensen, Križanauskienė, Lima, de Lope, Magnussen, Martin, Møller, Palinauskas, Pap, Pérez-Tris, Sehgal, Soler, Szöllősi, Westerdahl, Zetindjiev and Bensch2011) has been conducted since the development of molecular methods to study avian malaria. More detailed studies on avian malaria are important, as the migratory birds might carry parasites across international boundaries and potentially lead to significant health impacts and mortality of both wild and domestic birds (Žiegytė et al., Reference Žiegytė, Bernotienė, Bukauskaitė, Palinauskas, Iezhova and Valkiūnas2014; Valkiūnas et al., Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015, Reference Valkiūnas, Ilgūnas, Bukauskaite, Palinauskas, Bernotiene and Iezhova2017; Schoener et al., Reference Schoener, Harl, Himmel, Fragner, Weissenböck and Fuehrer2019).
To investigate vector–parasite relationships of avian Plasmodium spp., mosquito infectivity, laboratory experiments must be conducted with parasites obtained from natural infections. Polymerase chain reaction (PCR) is a highly sensitive tool to detect DNA from parasites in both avian hosts and mosquito vectors, but microscopic detection of sporozoites remains necessary to verify sporogony and to identify competent vectors (Valkiūnas, Reference Valkiūnas2011). However, the morphology of life stages within the vector (oocysts and sporozoites) does not vary sufficiently across Plasmodium species to allow for the identification of parasite species or lineage by microscopy (Santiago-Alarcon et al., Reference Santiago-Alarcon, Palinauskas and Schaefer2012), and therefore a combination of morphological and PCR studies is necessary (Valkiūnas, Reference Valkiūnas2011). When possible, the morphology of the parasite within erythrocytes (trophozoites, meronts and gametocytes) following experimental infection of an avian host is valuable in order to confirm the parasite morphospecies (Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018). This combined approach has been adopted in studies of human malaria parasites and has been increasingly employed to study avian malarial parasites (Kim and Tsuda, Reference Kim and Tsuda2010; Kazlauskienė et al., Reference Kazlauskienė, Bernotienė, Palinauskas, Iezhova and Valkiūnas2013; Žiegytė et al., Reference Žiegytė, Bernotienė, Bukauskaitė, Palinauskas, Iezhova and Valkiūnas2014; Valkiūnas et al., Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015).
In this study, we characterize Plasmodium lineage PADOM02 from naturally infected free-ranging sparrows (P. domesticus) in Giza, Egypt, including (i) morphological characterization of erythrocytic stages in the natural host, (ii) morphological characterization of the complete sporogony in Cx. pipiens and (iii) molecular characterization and phylogenetic analysis of the cytochrome b gene.
Material and methods
Detection and characterization of natural infection in sparrows
Ethics approval was obtained from the Institutional Animal Care and Use Committee of the University of Cairo for holding, transportation and laboratory maintenance of wild sparrows (CU-IF 89-19). Two hundred house sparrows (P. domesticus) were captured from Elsaf city, Giza Governorate, with traps and nets. The sparrows were brought to the laboratory alive and identified. A drop of blood was collected from the brachial vein of each bird to prepare blood smears, which were air-dried, fixed with methanol and stained with Giemsa. Approximately 100–150 fields were examined at low magnification (400X), and then at least 100 fields were studied at high magnification (1,000X), corresponding to a minimum 5 × 105 erythrocytes. Infection intensity, as a percentage, was estimated by counting the number of parasites per 1000 erythrocytes or per 10 000 erythrocytes if infections were very light (Godfrey et al., Reference Godfrey, Fedynich and Pence1987). Malaria parasites were identified following the keys provided by Valkiūnas (Reference Valkiūnas2005) and Valkiūnas and Iezhova (Reference Valkiūnas and Iezhova2018). Birds with mixed infections were excluded.
Experimental infection and sporogony characterization in Cx. pipiens complex
Mosquito larvae were obtained from breeding habitats in Giza Governorate, Egypt. Larvae were reared in the insectaries of the Department of Entomology, Faculty of Science, Cairo University, to obtain adults for morphological identification based on keys provided by Harbach (Reference Harbach1985). A stock colony of adult Cx. pipiens complex was maintained at 27 ± 2°C and 60–70% relative humidity (RH), following methods described by Adham et al. (Reference Adham, Gabre, Ayaad and Galal2003).
Approximately 50 unfed Cx. pipiens complex adult females were sexed and separated in experimental cages (25 × 25 × 25 cm3). These females were deprived of a sugar solution and then were allowed to feed fully on sparrows with single natural infection of Plasmodium PADOM02, whose blood contained mature gametocytes (0.1–0.2 parasite per erythrocyte), as described by Valkiūnas et al. (Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015). Single infected sparrow has been used for each replicate. Non-infected sparrows (as determined through blood smears) were used as controls. Fully fed female mosquitoes were separated in small cages with an aspirator and kept at 28°C for sporogony. A subset of control and infected females were removed and dissected as described by Valkiūnas (Reference Valkiūnas2005), at various intervals [(12 h, 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 8 days, 9 days and 10 days post infection (dpi)] to investigate parasite development. Infected and control Culex midgut contents were stained by Giemsa stain, in the same way as blood smear (Valkiūnas, Reference Valkiūnas2005), for ookinete visualization. Midgut permanent preparations were checked under binocular at 20X magnification for oocysts presence, while ookinetes and sporozoites have been examined at higher magnification. Histological preparations of midgut were prepared from infected and control females at the same time intervals then stained by hematoxylin and eosin to observe oocysts maturation. Finally, salivary glands and the residuals of the whole body (carcasses) were squashed on a slide to prepare a thick film then air-dried, fixed with methanol and stained by Giemsa (Valkiūnas et al., Reference Valkiūnas, Kazlauskiene, Bernotiene, Palinauskas and Iezhova2013).
DNA extraction, PCR and sequencing
Plasmodium parasites were identified within 24 h by microscopic examination for sparrow blood smears and all the positive ones were used for genomic DNA extraction and PCR amplification as follows: approximately 50 μL whole blood of the infected sparrows were drawn from the brachial vein into heparinized microcapillaries and stored in SET-buffer (0.015 M NaCl, 0.05 M Tris, 0.25 M EDTA, pH 8.0) and stored at −20°C for molecular analysis (Hellgren et al., Reference Hellgren, Waldenström, Peréz-Tris, Ösi, Hasselquist, Krizanauskiene, Ottosson and Bensch2007). The standard ammonium-acetate protocol was used for genomic DNA extraction from avian blood and Culex whole body after 4 and 6 days of infection (Sambrook et al., Reference Sambrook, Fritsch and Maniatis1989). Total DNA was quantified by BioPhotometer 6131 (Eppendorf, Hamburg, Germany) and analyzed on a 1% denatured agarose gel to ensure its integrity; the 260/280 and 260/230 absorbance ratios were evaluated for protein and solvent contamination. A nested-PCR protocol was used to amplify a 480-bp fragment of the mitochondrial cytochrome b (cyt-b) gene of Haemoproteus and Plasmodium, with primers HaemNF and HaemNR2 followed by primers HaemF and HaemR2 (Waldenström et al., Reference Waldenström, Bensch, Hasselquist and Östman2004). Amplifications were conducted with a 25 μL volume, with 2 μL of the first amplification product as a template for the second reaction. Amplification products were visualized on a 1% agarose gel stained with Ethidium bromide and photographed with a high-resolution gel documentation system. Uninfected house sparrow, which proved to be parasite free by microscopic examination, was used as a negative control.
Partial sequencing of cytochrome b gene and phylogenetic analysis
Amplification bands were excised from agarose gel and purified by Wizard SV Gel and PCR Clean-Up System kit (Promega, Madison, USA). Sanger sequencing was performed with HaemF and HaemR2 primers, with BigDye Terminator v3.1 and ABI PRISM 3100 (Applied Biosystems, Austin, USA). Resulting sequences were aligned and edited with BioEdit (Hall, Reference Hall1999), and deposited in Genbank (accession code MK018109).
Phylogenetic analysis of the cyt-b gene sequences was conducted to compare sequences obtained in this study to publicly available sequences of avian haemosporidians from public databases (Table S1). Only the 479 bp segment was used in this analysis, as standardized in the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009). Sequences were aligned by ClustalW (Thompson et al., Reference Thompson, Gibson and Higgins2002), as implemented in MEGA 6.06 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). Bayesian phylogenetic trees were produced with MrBayes 3.2.6 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003), based on the GTR + I + G model of nucleotide evolution recommended by jModelTest 2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). Two Markov chains for 10 million generations with sampling every 1000 generations; the first 2500 trees (25%) were discarded as a burn-in step and the remaining trees were used to calculate the posterior probabilities. Pairwise estimates of evolutionary distance were produced with MEGA 6.06 based on a maximum composite likelihood model, with a gamma distribution (shape parameter = 1), including transitions and transversions, and excluding ambiguous positions for each sequence pair.
Results
Morphological identification of Plasmodium in the avian host
Plasmodium parasite has been identified from 53 house sparrow blood smears (n = 200) with an overall prevalence of 26.5%. Only 3 mixed infections have been detected (1.5%) and have been excluded. The ones used in the transfection experiments were identified as Plasmodium (Haemamoeba) cathemerium based on the morphological characteristics seen in blood smears from the avian host (Fig. 1) and later confirmed by PCR reaction and sequencing of Cyt b fragment. Trophozoites are present in polychromatic (Fig. 1A and B) and mature erythrocytes (Fig. 1C and D). They have an irregular shape and uneven outline, seldom touch the host cell nucleus and do not displace or only slightly displace it (Fig. 1E and F). Erythrocytic meronts are large (but do not exceed 10 μ m in length) and usually roundish with abundant cytoplasm, markedly distorting the host cell and displacing the host cell nucleus (Fig. 1G, H and J) or occasionally enucleating (Fig. 1I and K) or rupturing the host cell (Fig. 1L). Pigmented granules in erythrocytic meronts are usually clumped into a spot, often near the margin of the parasite, and the number of merozoites ranges from 9 to 18 (most frequently, between 11 and 14). Macrogametocytes (Fig. 1M–P) and microgametocytes (Fig. 1Q–T) are generally roundish, markedly exceed the host cell nucleus, but neither surpass 10 μ m in length nor occupy the entire cytoplasm of the host cell. Gametocytes, especially microgametocytes, frequently enucleate the host cell (Fig. 1P and R–T). Pigmented granules are randomly scattered in the cytoplasm of gametocytes; however, rod-like pigmented granules with pointed ends are occasionally present in microgametocytes (Fig. 1S and T). Large vacuoles or residual bodies are absent in all parasites, and no meronts were observed outside of the blood cells.
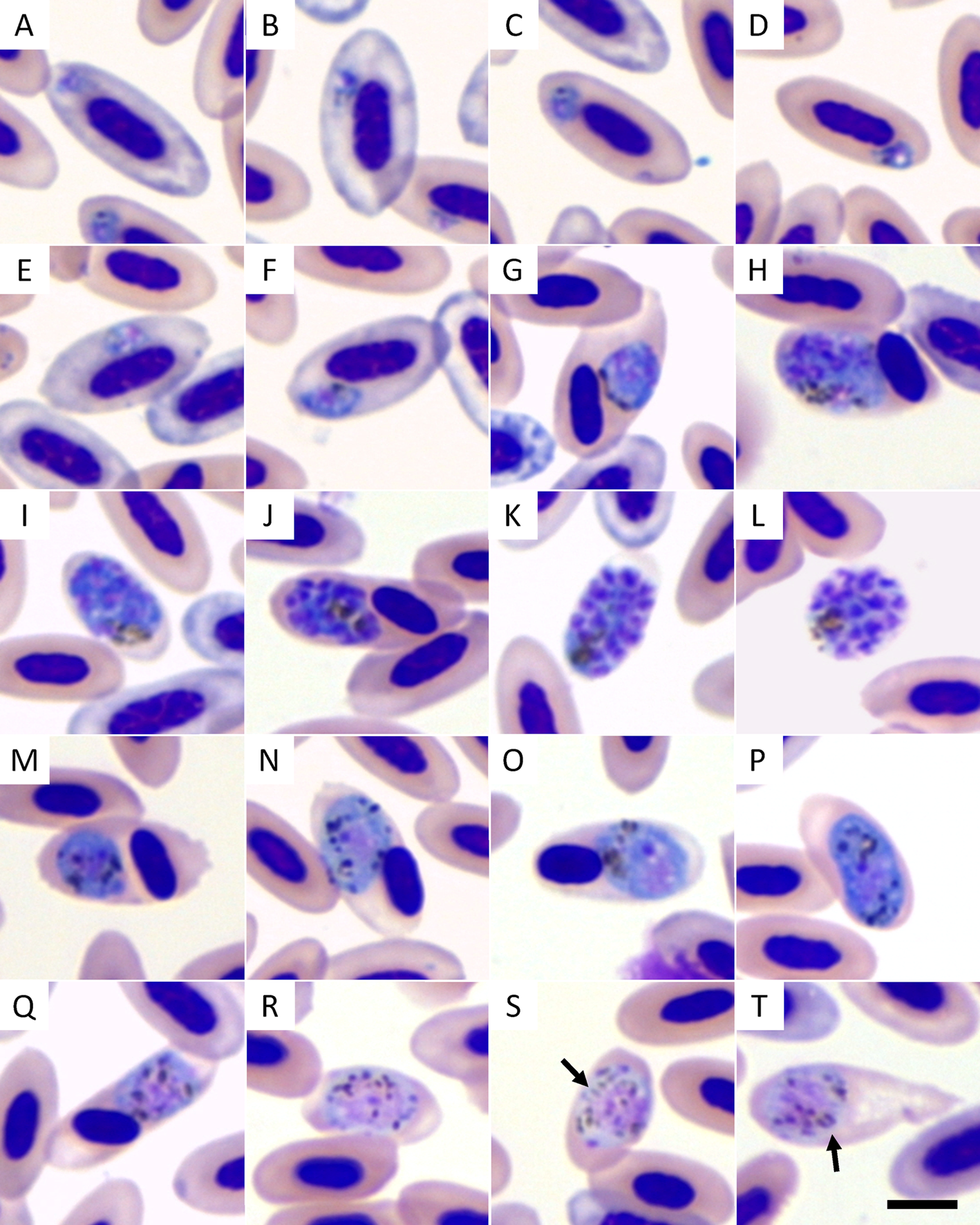
Fig. 1. Plasmodium cathemerium lineage PADOM02 a Giemsa-stained blood smears of a naturally infected sparrow (Passer domesticus). Legend: (A–F) trophozoites; (G–L) erythrocytic meronts; (M–P) macrogametocytes; (Q–T) microgametocytes. Arrows indicate rod-like pigmented granules. Scale bar = 5 μ m.
Parasite development in Cx. pipiens complex
Culex pipiens complex mosquitoes were susceptible to experimental infection after being allowed to feed on sparrows with a parasitemia of 0.1 to 0.2 parasites per erythrocyte and gave positive PCR results. Ookinetes were seen in the midgut of Cx. pipiens complex as early as 24 h after exposure; these structures were large curved-elongated bodies (Fig. 2A, Table 1). The nuclei of ookinetes had an irregular outline and were positioned slightly laterally, and numerous dark granules were scattered in the deep-blue cytoplasm (Fig. 2A). The number of ookinetes decreased rapidly, and only very few ookinetes were seen in the midgut of mosquitoes examined 2 dpi.
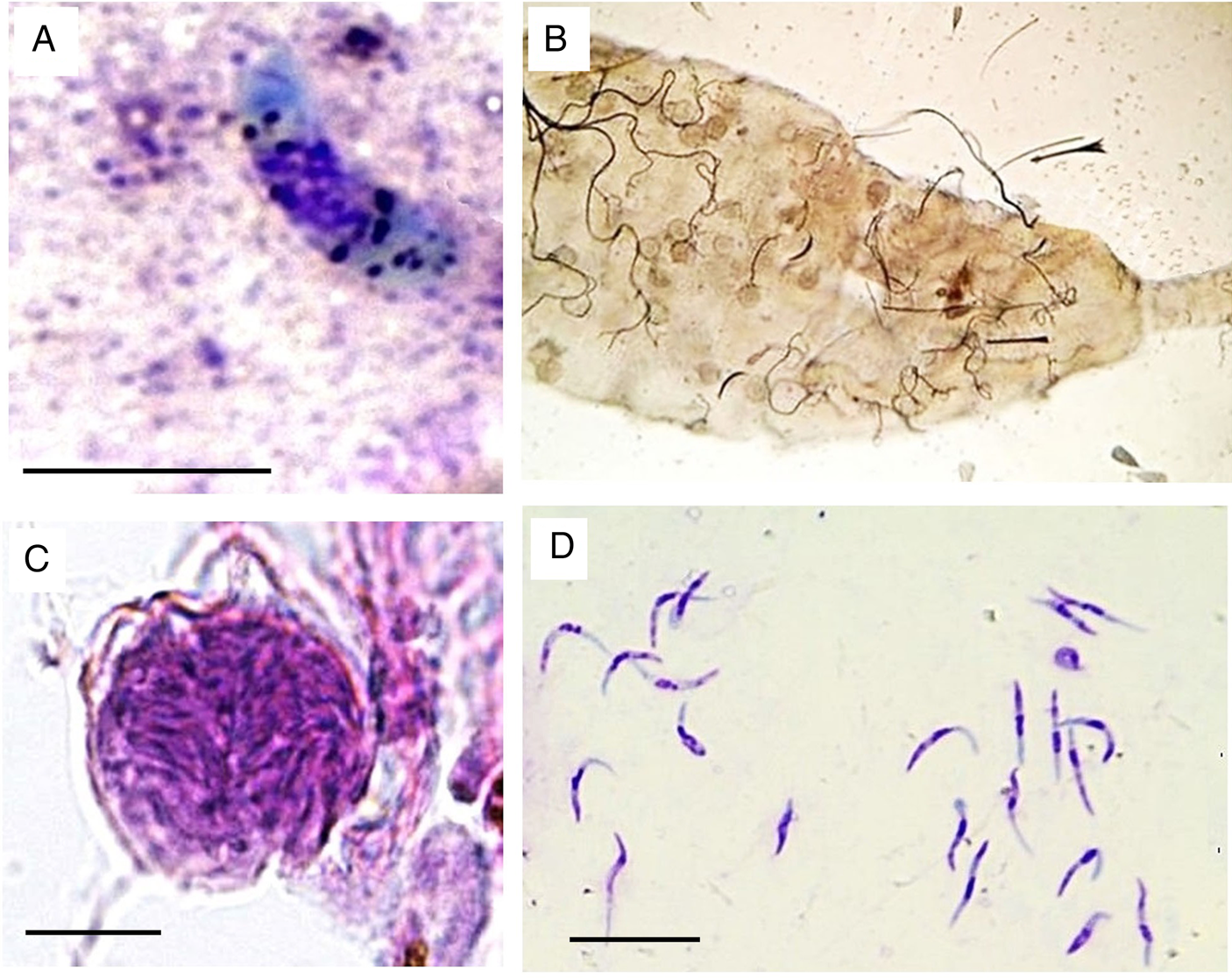
Fig. 2. Plasmodium cathemerium lineage PADOM02 in the tissues of experimentally-infected Culex pipiens complex: (A) mature ookinete in Giemsa-stained midgut contents preparation obtained 1 day post infection (dpi) (B) mosquito midgut inundated by oocyst as observed under light microscope (20X) (C) mature oocysts in hematoxylin and eosin-stained midgut histological preparations obtained 4 dpi (D) mature sporozoites in Giemsa-stained salivary gland preparations obtained 5 dpi. Scale bar = 10 μ m.
Table 1. Morphometry of Plasmodium cathemerium lineage PADOM02 in Culex pipiens complex

a Methanol-fixed preparations at 1 day post infection (dpi).
b Formalin-fixed preparations of mature parasites at 4 dpi.
c Methanol-fixed preparations at 5 dpi.
Oocysts developed rapidly but asynchronously and were seen in large numbers (≥54) in the midgut of mosquitoes examined as early as 4 dpi (Fig. 2B). Maturing oocysts varied considerably in size (Table 1), containing 9 to 19 pigment granules scattered in their cytoplasm as well as a prominent vacuole (Fig. 2C). Development continued, leading to the formation of sporoblasts and the release of sporozoites. The fusiform sporozoites were first seen in salivary glands 6 dpi and continued to be present until the end of the experiment (10 dpi). Thousands of sporozoites were seen in preparations of salivary glands, indicating successful sporogony. The sporozoites were curved-bodied, with blunt ends and possessed centrally located nuclei (Fig. 2D, Table 1).
PCR, sequencing and phylogenetic analysis
Sequencing of the cytochrome b amplified fragments from both donor sparrow blood and infected female mosquitoes in 4 and 6 days after infection (when mature oocysts and sporozoites stages morphologically detected) produced a 480 bp partial sequence that was 100% identical to several published sequences (Genbank accession codes AB474380, AB474381, AB542062, DQ659543, DQ838991, HF543659 and KJ488599). When only the sequence trimmed as standardized by MalAvi (479 bp) was evaluated, the parasite was identical to lineage PADOM02 (Bensch, Reference Bensch2019). Phylogenetic analysis revealed that the cyt-b gene sequence of PADOM02 clusters with lineages SEIAUR01 and PADOM09, both of which have been attributed to the morphospecies Plasmodium cathemerium. The evolutionary distance of the cyt-b gene sequence of PADOM02 was smaller in relation to other lineages attributed to P. cathemerium (0.63% – 1.50% expected base substitutions per site) than to lineages attributed to other species of the Haemamoeba subgenus (3.08–5.74%) (Fig. 3Table 2)

Fig. 3. Bayesian phylogenetic tree of the mitochondrial cytochrome b gene sequences (480 bp) of the studied haemosporidian lineage. MalAvi or GenBank accession codes for each lineage are indicated within brackets. Branch lengths are drawn proportionally to evolutionary distance. Values adjacent to nodes correspond to posterior probabilities.
Table 2. Estimates of evolutionary distance (% expected base substitutions per site) of mitochondrial cytochrome b gene sequences of Plasmodium PADOM02 in relation to lineages attributed to the Haemamoeba subgenus

Discussion
Although Africa has a remarkable avian diversity, with ~2700 species (Avibase – The World Bird Database https://avibase.bsc-eoc.org/avibase.jsp?lang=EN), the diversity of avian malarial parasites in this continent has been considerably understudied (Valkiūnas, Reference Valkiūnas2005). Many wild bird species, especially migratory birds, are susceptible to various malarial infections (Clark et al., Reference Clark, Clegg and Lima2014). Only a few studies have evaluated the occurrence of Plasmodium spp. in Egyptian birds (Haiba, Reference Haiba1948; Mohammed, Reference Mohammed1958a, Reference Mohammedb; Guindy et al., Reference Guindy, Hoogstraal and Mohammed1965; Wiersch et al., Reference Wiersch, Maier and Kampen2005); however, no study has been done on their vectors. The study of Marzal et al. (Reference Marzal, Ricklefs, Valkiūnas, Albayrak, Arriero, Bonneaud, Czirják, Ewen, Hellgren, Hořáková, Iezhova, Jensen, Križanauskienė, Lima, de Lope, Magnussen, Martin, Møller, Palinauskas, Pap, Pérez-Tris, Sehgal, Soler, Szöllősi, Westerdahl, Zetindjiev and Bensch2011) has detected 3 Plasmodium lineages PADOM02, PADOM16 and SGS1 in house sparrows from Luxor (south part of Egypt) with 85.3% prevalence and 2.9% mixed infection. The current study record a less prevalence per cent, 26.5% in P. domesticus captured from Giza Governorate, Egypt with 1.5% mixed infection. The data recorded in Giza is lower than previously detected in Luxor suggested the effect of urbanization on both bird and vector (Martínez-De la Puente et al., Reference Martínez-De la Puente, Ferraguti, Ruiz, Roiz, Soriguer and Figuerola2016; Carbo-Ramirez et al., Reference Carbo-Ramirez, Zuria, Schaefer and Santiago-Alarcon2017). Additionally, we morphologically characterized the erythrocytic stages inside the erythrocytes of the avian host and the sporogonic cycle in the vector, combined with a phylogenetic analysis of mitochondrial cyt-b gene. The lineage PADOM02 has been detected in a number of avian hosts and mosquito vectors worldwide (Table 1). Most studies reporting this lineage have detected it solely through molecular methods, and therefore there is limited information on its morphology. Beadell et al. (Reference Beadell, Ishtiaq, Covas, Melo, Warren, Atkinson, Bensch, Graves, Jhala, Peirce, Rahmani, Fonseca and Fleischer2006) originally attributed this lineage to the morphospecies Plasmodium (Haemamoeba) relictum. However, this classification must be considered with caution, because that study did not provide details on the morphological criteria that were used. Furthermore, later studies have found that another sequence attributed to P. relictum in the study of Beadell et al. (Reference Beadell, Ishtiaq, Covas, Melo, Warren, Atkinson, Bensch, Graves, Jhala, Peirce, Rahmani, Fonseca and Fleischer2006) corresponded to Plasmodium (Huffia) elongatum (Valkiūnas et al., Reference Valkiūnas, Zehtindjiev, Dimitrov, Križanauskienė, Iezhova and Bensch2008; Vanstreels et al., Reference Vanstreels, Kolesnikovas, Sandri, Silveira, Belo, Junior, Epiphanio, Steindel, Braga and Catão-Dias2014). When D'Amico and Baker (Reference D'Amico and Baker2010) examined the blood smear of a red knot (Calidris canutus) infected with this lineage, they concluded that it corresponded to P. relictum based on the classification that had been proposed by Beadell et al. (Reference Beadell, Ishtiaq, Covas, Melo, Warren, Atkinson, Bensch, Graves, Jhala, Peirce, Rahmani, Fonseca and Fleischer2006). D'Amico and Baker (Reference D'Amico and Baker2010) did not provide a detailed morphological description, however, the basic text description and the photographs provided in their study are generally consistent with several species of the subgenus Haemamoeba.
The parasites in our study have roundish gametocytes that are markedly larger than the host cell nucleus, which unequivocally places them in the subgenus Haemamoeba, with 11 known species (Valkiūnas, Reference Valkiūnas2005; Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018). The absence of large vacuoles or residuals bodies in the trophozoites and erythrocytic meronts provides the basis for the elimination of P. giovannolai, P. matutinum, P. tejerai and P. griffithsi (Valkiūnas, Reference Valkiūnas2005; Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018). Because the erythrocytic meronts do not cause a marked enlargement of the host cell, do not possess a centrally located residual cytoplasm, and their nuclei are not located in the periphery, P. caloti and P. parvulum can be excluded (Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018). Because fully grown gametocytes neither exceed 10 μ m in length nor occupy the entire cytoplasm of the host cell, and the gametocyte pigmented granules are not clumped into a spot, it is also possible to exclude P. gallinaceum, Plasmodium coturnixi, and P. lutzi (Valkiūnas, Reference Valkiūnas2005; Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018). There are thus three morphospecies within the subgenus Haemamoeba to be further considered: P. cathemerium, P. relictum and P. subpraecox. Of these, we consider our parasite to be morphologically most consistent with P. cathemerium based on the following characteristics: (a) presence of elongated rod-like pigmented granules, with pointed ends in the microgametocytes (a characteristic thought to be unique to P. cathemerium); (b) when a single trophozoite is present, the host cell nucleus is only occasionally and slightly displaced (unlike the high frequency and marked nuclear displacement that is usually associated with P. relictum); (c) 9–18 merozoites per erythrocytic meronts is consistent with the interval of 6–24 merozoites described for P. cathemerium (whereas P. subpraecox usually has 5–12 merozoites); (d) the infection was detected in a house sparrow (whereas previous studies have shown this species to be refractory to P. subpraecox) (Valkiūnas, Reference Valkiūnas2005; Valkiūnas and Iezhova, Reference Valkiūnas and Iezhova2018). We, therefore, conclude that the original assignment of the lineage PADOM02 to the morphospecies P. relictum was in error and that this lineage is instead most consistent with P. cathemerium. This interpretation is corroborated by our phylogenetic analysis, which places PADOM02 amidst other lineages attributed to P. cathemerium.
Culex pipiens is the most common vector of avian malaria (Valkiūnas, Reference Valkiūnas2005; Santiago-Alarcon et al., Reference Santiago-Alarcon, Palinauskas and Schaefer2012). Huff (Reference Huff1927, Reference Huff1932) found that the following mosquitoes are susceptible to P. cathemerium (unknown cyt-b lineage): Cx. pipiens, Cx. salinarius, Cx. territans, Cx. quinquefasciatus, Cx. tarsalis and Aedes aegypti. However, Huff (Reference Huff1927) has reported the transfer from birds to mosquitoes and from mosquitoes back to birds with P. cathemerium with only two species of the studied mosquitoes. A recent study with morphological and molecular methods to detect Plasmodium spp. in Cx. pipiens from Austria has identified three lineages: P. vaughani SYAT05, P. elongatum GRW6 and P. relictum SGS1 (Schoener et al., Reference Schoener, Harl, Himmel, Fragner, Weissenböck and Fuehrer2019). In this study, we found that Cx. pipiens complex (adults derived from larvae collected in Egypt) were susceptible to P. cathemerium PADOM02. The sporogonic process and the morphology of sporogonic stages of P. cathemerium PADOM02 in Cx. pipiens complex were generally comparable to those of P. relictum GRW4 in Cx. pipiens pipiens (Valkiūnas et al., Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015), and it seems unlikely that the vector stages of these parasites could be differentiated on a morphological basis. The massive infection of the salivary glands suggests that a substantial number of sporozoites would be inoculated through blood-sucking, corroborating that Cx. pipiens complex is a competent vector of PADOM02. In contrast with Valkiūnas et al. (Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015), however, we observed a much faster developmental rate, with oocysts developing in 4 dpi and sporozoites in 5 dpi (cf. 13 dpi for oocysts and 16 dpi for sporozoites in Valkiūnas et al., Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015). This is likely related to the higher ambient temperatures used to rear mosquitoes in this study (27 ± 2°C, compared to 20 ± 1°C in Valkiūnas et al., Reference Valkiūnas, Žiegytė, Palinauskas, Bernotienė, Bukauskaitė, Ilgūnas, Dimitrov and Iezhova2015) as temperature effects on the Plasmodium extrinsic incubation period (the interval of plasmodium development inside the vector from blood meal acquisition to salivary gland infection). The complete incubation period lasts 5–16 days, is inversely dependent on temperature and ultimately restricts the altitudinal distribution of infectious mosquitoes and disease transmission (LaPointe et al., Reference LaPointe, Goff and Atkinson2010, Reference LaPointe, Atkinson and Samuel2012; Carvalho et al., Reference Carvalho, Rangel and Vale2017).
The only other study employing molecular methods to identify the Plasmodium lineages in Egyptian birds was conducted by Marzal et al. (Reference Marzal, Ricklefs, Valkiūnas, Albayrak, Arriero, Bonneaud, Czirják, Ewen, Hellgren, Hořáková, Iezhova, Jensen, Križanauskienė, Lima, de Lope, Magnussen, Martin, Møller, Palinauskas, Pap, Pérez-Tris, Sehgal, Soler, Szöllősi, Westerdahl, Zetindjiev and Bensch2011). Interestingly, in that study, Plasmodium lineage PADOM02 was also detected in free-ranging sparrows despite a relatively small sample size (34 individuals). The fact that we detected the same parasite in sparrows captured ~500 km north from the study site of Marzal et al. (Reference Marzal, Ricklefs, Valkiūnas, Albayrak, Arriero, Bonneaud, Czirják, Ewen, Hellgren, Hořáková, Iezhova, Jensen, Križanauskienė, Lima, de Lope, Magnussen, Martin, Møller, Palinauskas, Pap, Pérez-Tris, Sehgal, Soler, Szöllősi, Westerdahl, Zetindjiev and Bensch2011) suggests that this is a common parasite in free-ranging sparrows along the Nile river. This is not unexpected considering the broad distribution of this lineage, which suggests this is a relatively common parasite of birds in the Holarctic region (see Table 3). Considering that the Nile river is an important stop-over site for migratory birds along the Black Sea-Mediterranean and East Africa-West Asia fly ways (Davidson and Stroud, Reference Davidson, Stroud, Boere, Galbraith and Stroud2006; Abdelwhab and Hafez, Reference Abdelwhab and Hafez2011), this raises interesting questions on the potential host of sparrows as a reservoir of infection for the transmission of these parasites to other birds passing through the area. It is also worth noting that the records of PADOM02 in yellowhammers (Emberiza citronella) in New Zealand (Bonneaud et al., Reference Bonneaud, Pérez-Tris, Federici, Chastel and Sorci2006; Ewen et al., Reference Ewen, Bensch, Blackburn, Bonneaud, Brown, Cassey, Clarke and Pérez-Tris2012) suggest that this parasite was introduced together with its host since yellowhammers are not native to New Zealand and were brought from Europe (Duncan, Reference Duncan1997).
Table 3. Summary of the known host–parasite associations of Plasmodium lineage PADOM02

a = Blood meal analyses revealed that the mosquitoes had recently fed on Corvus macrorhynchos, Passer montanus and Parus major (Kim et al., Reference Kim, Tsuda, Yamada and Yamada2009) or on Ardea cinerea, Strix uralensis and Bos taurus (Ejiri et al., Reference Ejiri, Sato, Kim, Hara, Tsuda, Imura, Murata and Yukawa2011).
b = Experimental infection from naturally infected Passer domesticus.
Although there are records of PADOM02 infecting Galliformes (Ishtiaq et al., Reference Ishtiaq, Gering, Rappole, Rahmani, Jhala, Dove, Milensky, Olson, Peirce and Fleischer2007) and Charadriiformes (D'Amico and Baker, Reference D'Amico and Baker2010), most records of this lineage thus far have been obtained from Passeriformes (see Table 3), suggesting these are the main hosts for this parasite. Lineages PADOM09 and SEIAUR01, which are also attributed to P. cathemerium, thus far have been predominantly reported in Passeriformes, with PADOM09 restricted to the Americas whereas SEIAUR01 appears to be restricted to North America and the Caribbean (Bensch, Reference Bensch2019). Further studies on the geographic and host distribution of avian malarial parasites at the lineage level will, therefore, provide valuable insight into the ecological dynamics of these parasites and their interactions with avian host. Molecular identification and evolutionary study provide a level of insight that could not be obtained exclusively by morphological studies since these parasites appear to be otherwise undistinguishable.
In conclusion, our findings confirm that Plasmodium lineage PADOM02 infects sparrows in urban areas along the Nile River, Egypt and corroborate that Cx. pipiens complex is a highly competent vector for these parasites. Furthermore, we demonstrate that this lineage corresponds to the morphospecies P. cathemerium and not P. relictum as previously believed.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182020000566.
Acknowledgements
We thank Prof. James Nation, Institute of Food and Agricultural Sciences, Entomology and Nematology Department, University of Florida, USA for his comments, valuable suggestions and English language editing. This work has been supported by Faculty of Science Research Sector in Cairo and South Valley Universities.
Financial support
This study was supported by research grand from Faculty of Science, Cairo University.
Conflict of interest
The authors have declared that they have no conflict of interest.
Ethical standards
This work was carried out with approval of Cairo University Institutional Animal Care and Use Committee (CU-IACUC), approval number CU-IF 89-19.









