Article contents
Molecular phylogenetic analysis of the genus Gyrodactylus (Platyhelminthes: Monogenea) inferred from rDNA ITS region: subgenera versus species groups
Published online by Cambridge University Press: 05 December 2003
Abstract
Analyses of small subunit ribosomal RNA gene sequences of representatives of major taxa of Monopisthocotylea were performed to identify the sister group of Gyrodactylus. Nuclear ribosomal DNA sequencesNucleotide sequence data reported in this paper are available in the GenBank, DDBJ and EMBL databases under the accession numbers AJ566375-79, AJ566768, AJ567670-74, AJ581657. from the complete internal transcribed spacer (ITS) region were used to infer phylogeny of 37 Gyrodactylus species and Gyrodactyloides bychowskii, Macrogyrodactylus polypteri and Gyrdicotylus gallieni, using maximum likelihood, parsimony and Bayesian inference. The genus Gyrodactylus appeared to be a monophyletic group in all analyses, based on the present data set. Within the genus, there were 3 major groups recognized by high bootstrap values and posterior probabilities. None of the 6 subgenera appeared to be monophyletic, and the most basal subgenus G. (Gyrodactylus) was paraphyletic. Characteristics of the excretory system of Gyrodactylus do not seem to be conservative enough to reveal subgenera within Gyrodactylus and we suggest abandoning existing subgenera as indicators of phylogeny. The grouping of species based on the morphology of the ventral bar and marginal hooks seems to have sufficient power to infer relationships between the Gyrodactylus species.
Keywords
- Type
- Research Article
- Information
- Copyright
- 2003 Cambridge University Press
INTRODUCTION
Members of the family Gyrodactylidae (Monogenea: Monopisthocotylea) are flatworm parasites with a large range of host organisms, predominantly teleost fish (William & Jones, 1994). Within the Gyrodactylidae, Gyrodactylus is the most diverse and widespread genus.
Based on the morphology of the excretory system, 6 Gyrodactylus subgenera were proposed by Malmberg (1956, 1964); G. (Gyrodactylus), G. (Mesonephrotus), G. (Metanephrotus), G. (Paranephrotus), G. (Neonephrotus), and G. (Limnonephrotus). Difficulties in the use of excretory system characters for systematic studies have arisen because the components of the protonephridial system are only clearly visible in live parasites, rendering fixed specimens all but useless for differentiating subgenera. For this reason, not all described Gyrodactylus species may be readily assigned to subgenera. The shape of the marginal hook was proven to be another character suitable to group Gyrodactylus species. Using this character, groups such as the G. elegans- or G. wageneri-group were established (see Malmberg, 1964, 1970 for more details) including other species of similar marginal hook morphology.
Due to advances in molecular biology, genetic markers for species identification of gyrodactylids have been investigated, based mainly on the ribosomal RNA (rRNA) genes and the associated internal transcribed spacers (ITS) (Cunningham et al. 1995 a; Cunningham, 1997). The ITS sequences from approximately 55 described Gyrodactylus species are known (Cunningham, 1997; Cable et al. 1999; Zietara et al. 2000; Cunningham et al. 2001; Matejusová et al. 2001; Huyse & Volckaert, 2002; Zietara & Lumme, 2002) with other sequences obtained from as-yet unidentified species (Matejusová & Cunningham, unpublished data; Zietara & Lumme, 2002).
Although members of the genus Gyrodactylus show high species diversity, with 402 valid species descriptions (Bakke, Harris & Cable, 2002), there have been no analyses that included exemplar species of all defined subgenera in order to display their phylogenetic relationship or to test their proposed monophyly. A phylogenetic analysis inferred from the combined 5.8S and ITS2 sequences of 10 gyrodactylids has given us partial information about the relationships among 3 subgenera and demonstrated a separation of G. (Limnonephrotus) from G. (Mesonephrotus) and G. (Metanephrotus) (Cable et al. 1999). Also, certain species belonging to the subgenus G. (Gyrodactylus) were suggested to be distant from species of the subgenus G. (Limnonephrotus) based on differences in the ITS1 and the morphology of haptor structures, in particular the marginal hooks and ventral bar (Matejusová et al. 2001). Subsequently, using 10 Gyrodactylus species belonging to 4 subgenera, maximum likelihood analysis inferred from the 5.8S sequences presented a monophyletic origin of Gyrodactylus with G. (Mesonephrotus) and G. (Metanephrotus) being a sister group to G. (Paranephrotus) and G. (Limnonephrotus) species (Zietara et al. 2002). The same authors also demonstrated that each of the subgenera possessed a unique 5.8S gene sequence.
Here we present phylogenetic analyses inferred from the ITS sequences of members of all Gyrodactylus subgenera to elucidate their relationships. Prior to this analysis, sequences of the small subunit (SSU) ribosomal RNA (rRNA) gene were used to identify a sister group of Gyrodactylus. SSU rRNA has been used successfully for determining the interrelationships of monopisthocotylean monogeneans and a substantial database is now available (e.g. Olson & Littlewood, 2002).
MATERIALS AND METHODS
Sequence alignment
The small subunit (SSU) ribosomal RNA gene of 6 Gyrodactylus species, Gyrodactyloides bychowskii and Macrogyrodactylus polypteri were sequenced. DNA extraction and PCR was carried out according to Matejusová et al. (2001), the primers used to amplify the SSU region were as described by Cunningham et al. (1995 b).
SSU sequences together with sequences of the complete internal transcribed spacers (ITS1 and ITS2) and 5.8S ribosomal DNA of 37 Gyrodactylus species, G. bychowskii, Gyrdicotylus gallieni Vercammen Grandjean, 1960 (AJ001843) and M. polypteri were aligned in CLUSTAL X (Jeanmougin et al. 1998), using default parameters. The full list of taxa used in the SSU and ITS rDNA alignments are shown in Tables 1 and 2 respectively, with GenBank accession numbers and specimen details for new sequences. The extraction, PCR, and sequencing methods for the ITS region of newly sequenced species were as described by Matejusová et al. (2001). Alignments were refined by eye using MacClade v. 4.05 (Maddison & Maddison, 2000). Regions of ambiguity were recorded and approximately 400 bp from the 5′ end of ITS1 were removed prior to analysis. Although the majority of positions were alignable among all taxa, it was difficult to satisfactorily align some ITS positions. All analyses were carried out using only positions that were unambiguously alignable across all taxa.


New SSU sequences were aligned to the monopisthocotylean portion of an existing published alignment of the monogenean SSU (Olson & Littlewood, 2002; EBI accession ALIGN_000146; see Table 1 for list of taxa), using the profile alignment option. The SSU analyses were performed to identify the most basal taxon of Gyrodactylus that would be used to root final trees of Gyrodactylus spp. based on the complete ITS sequences.
The full alignments for the SSU and ITS data sets (21 and 40 species respectively) have been deposited with EBI and are available by anonymous FTP from ftp.ebi.ac.uk in directory /pub/databases/embl/align and via the EMBLALIGN database via SRS at http://srs.ebi.ac.uk, under the following accessions ALIGN_000604 (SSU) and ALIGN_000605 (ITS). Exclusion sets are added as notes and the alignments may be adapted as NEXUS files.
Phylogenetic analyses
We estimated phylogenies using maximum parsimony (MP), Bayesian inference (BI) and maximum likelihood (ML), rooting the ingroup against Anoplodiscus cirrusspiralis for the SSU data set. Following the SSU analyses, ITS phylogenies were rooted against Gyrodactyloides, Macrogyrodactylus and Gyrdicotylus.
MP and ML analyses were conducted with PAUP*4.0b10 (Swofford, 2002), employing a branch-and-bound search strategy for MP and a heuristic search strategy for ML. Modeltest v. 3.06 was used to select the model of evolution of best fit for each data partition (Posada & Crandall, 1998). For the SSU data and for each of the ITS1, 5.8S and ITS2 partitions individually, we employed a GTR+I+G model; this refers to a general-time-reversible model including estimates of invariant sites and gamma distributed among-site rate variation. BI was determined using MrBayes (Huelsenbeck & Ronquist, 2001, ver. 2.01) with the following parameters: nst=6, rates=invgamma, ncat=4, shape=estimate, inferrates=yes, and basefreq=empirical, that corresponds to a GTR+I+G substitution model. For the ITS data, each of the data partitions were treated independently, and each using an independently estimated GTR+I+G substitution model. Posterior probabilities were approximated over 1000000 generations (ngen=1000000) via 4 simultaneous Markov Chain Monte Carlo chains (MCMC) (nchains=4) with every 100th tree saved (samplefreq=100). Default values were used for the MCMC parameters. Consensus trees with mean branch lengths were constructed using the ‘sumt’ command with the ‘contype=allcompat’ option and ignoring the initial topologies saved during ‘burn in’; the initial n-generations before log-likelihood values and substitution parameters plateau (see Huelsenbeck & Ronquist, 2001). MP and ML nodal supports were estimated by bootstrap analyses (heuristic search, 1000 replicates for MP, 100 replicates for ML), and as posterior probabilities in the Bayesian inference analyses (Huelsenbeck et al. 2001).
RESULTS
SSU rDNA
The new SSU sequence length varied from 1892 bp (G. rhodei) to 1974 bp (G. sedelnikowi). The complete alignment spanned 2189 positions but only 1579 were included and considered unambiguously aligned. Of these, 1117 positions were constant and 323 informative under the principles of parsimony. Modeltest found that the most appropriate model of substitution was GTR+I+G and we used this for both ML and BI. For ML the following parameters were used: rate matrix, 0·9654 (A–C), 4·0010 (A–G), 2·3835 (A–T), 0·8694 (C–G), 5·4764 (C–T), 1·0000 (G–T); nucleotide frequencies A=0·2713, C=0·2069, G=0·2612, T=0·2606; assumed proportion of invariable sites=0·4782; gamma shape parameter (alpha)=0·6441; 4 rate categories. ML and BI analyses performed on the SSU data set resolved trees with an identical topology, and almost identical (relative) branch lengths. MP found 6 equally parsimonious trees (length=1019; CI=0·618; RI=0·741; RC=0·458) and the strict consensus was also fully compatible with the single tree topology inferred by ML and BI. The ML tree is shown in Fig. 1, with branch lengths estimated by ML and nodal support from ML (bootstrap, n=100), MP (bootstrap, n=1000) and BI (posterior probabilities).

Fig. 1. Phylogeny of monopisthocotylean Monogenea based on SSU rDNA indicating the relative position of Gyrodactylus species and potential outgroup taxa. The tree topology is from a maximum likelihood analysis with nodal support indicated, from top to bottom, for maximum likelihood (bootstrap %, n=100), maximum parsimony (bootstrap %, n=1000) and Bayesian inference (posterior probabilities); see text for further details.
The BI solution, a consensus of 2560 trees, further resolves G. rhodei+G. rutilensis and G. gobiensis+G. salaris (G. (Limnonephrotus) subgenus) as sister taxa of G. carassii+G. sedelnikowi (G. (Gyrodactylus) subgenus) with posterior probabilities of 100 in this sample. In addition, the genus Gyrodactylus appears monophyletic with M. polypteri as its sister group; G. bychowskii was resolved as the sister group to Gyrodactylus+Macrogyrodactylus.
ITS rDNA
MP analysis found 3 equally parsimonious trees (length=1357; CI=0·491; RC=0·338). An incongruence length difference test (Farris et al. 1994), as implemented in PAUP*, suggested that the individual data partitions had evolved significantly differently from one another (P=0·007) and were not combinable in a phylogenetic analysis with the same nucleotide substitution model. Modeltest found that the most appropriate model of substitution was GTR+I+G for each of the data partitions and when these partitions were combined. There was little difference in tree topology whether these partitions were modelled separately or combined. We used the model for both ML and BI. For BI, where each of the partitions was modelled separately, we estimated that log likelihood values had reached a plateau at approximately 40000 generations. We ignored results for a further 20000 generations and summarized trees for the final 940000 generations (9400 trees). Branch lengths were calculated as means of the branch lengths in the individual topologies saved during Bayesian analysis and summarized using the ‘sumt’ command of MrBayes.
All 3 analyses resolved the same broad patterns of evolution among the Gyrodactylus species. The only topological differences between the phylogenetic solutions were amongst poorly supported clades. We show only the solution derived from Bayesian analysis using the independent GTR+I+G estimates for each of the 3 combined data partitions, which was almost identical to that of BI and ML using a single model, with nodal support from BI and MP (Fig. 2) where posterior probabilities exceed 80% and MP bootstrap exceed 50%; it was computationally impossible to provide bootstrap support for the ML analysis using the most appropriate model of substitution.
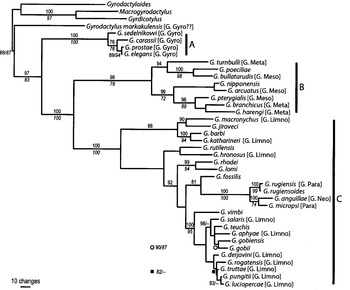
Fig. 2. Phylogeny of Gyrodactylus species based on ITS rDNA, rooted against Gyrodactyloides bychowskii. The tree topology is from a Bayesian analysis, modelling each data partition separately, with nodal support indicated, from top to bottom, for Bayesian inference (posterior probabilities) and maximum parsimony (bootstrap %, n=1000) where these values are >80% and 50% respectively. Subgenera, where known, are indicated in square brackets as: G. Gyro – G. Gyrodactylus; G. Limno – G. Limnonephrotus; G. Meso – G. Mesonephrotus; G. Meta – G. Metanephrotus; G. Neo – G. Neonephrotus; G. Para – G. Paranephrotus according to terms proposed by Malmberg (1956, 1970); see text for further details.
At the base of the Gyrodactylus clade, G. markakulensis was consistently resolved as the most basal taxon. Next, the remaining taxa within the subgenus G. (Gyrodactylus) were resolved to be strongly monophyletic, with each of the analyses resolving the same interrelationships as indicated in Fig. 2. The next clade to be resolved was a mixture of taxa in the subgenera G. (Metanephrotus) and G. (Mesonephrotus). BI and ML resolved identical topologies within this clade, but MP pulled G. turnbulli to the base. The third well-supported clade comprises the remaining taxa, in which some nodes were strong and others indicated poor resolution. BI and ML resolved almost identical topologies; differences concerned only the interrelationships of the most derived taxa where branch lengths were very short. However, within this group, MP resolved the G. rugiensis+G. rugiensoides, G. anguillae+G. micropsi clade as the most basal taxa, and G. rutilensis and G. hronosus as more derived and not as sister taxa; these differences in the MP analysis account for the low bootstrap proportions plotted at the nodes of the Bayesian tree (Fig. 2).
DISCUSSION
The present study analysed exemplar species of all defined subgenera of Gyrodactylus and brings more insights into the phylogenetic relationship within the genus. As the most commonly sequenced region of the genome, the complete ITS rDNA sequences of 37 Gyrodactylus species were used to infer the phylogeny. This region has proven valuable within Gyrodactylus (Cable et al. 1999; Zietara et al. 2002) and demonstrated the potential to resolve the phylogeny of different monogeneans (Bentz et al. 2001), parasite groups (e.g. Audebert, Durette-Desset & Chilton, 2000) or plants (Steane et al. 1999).
Three phylogenetic methods revealed trees of almost identical topology, with strong support at some critical nodes and confirmed the ability of the ITS to infer phylogeny in Gyrodactylus in our data set. Gyrodactylus falls into 3 well-supported clades, suggesting a basal, but not monophyletic, origin of the G. (Gyrodactylus) subgenus (G. markakulensis and the taxa in clade A). Separation of the G. (Gyrodactylus) species from the others is not surprising, considering the molecular and morphological data. Even in the relatively conserved SSU rDNA (V4 region) (see Cunningham et al. 1995 a), these species varied by up to 12% from those of the G. (Limnonephrotus) subgenus (Matejusová et al. 2001). The basal position of G. (Gyrodactylus) in the whole genus might be confirmed by the fact that this subgenus shares some plesiomorphic characters, such as the median junction between the two anterior systems of the excretory system, with the genus Macrogyrodactylus (see Malmberg, 1964, 1970), which has been considered the closest ancestor of Gyrodactylus (Malmberg, 1998). Based on the morphology of the attachment apparatus, mainly the ventral bar, some similarities can be drawn between the present members of the G. (Gyrodactylus) subgenus. Typically, the ventral bar has a long narrow membrane and no lateral processes (Gyrodactylus elegans-group) or a tongue-shape membrane with very short or no lateral processes (Gyrodactylus phoxini-group) (see Malmberg, 1970). The position of G. markakulensis is exceptional as it was consistently resolved as the most basal taxon in all phylogenetic analyses performed. However, the position of this species is controversial, and it was included, albeit with some reservations, in both G. elegans- and G. phoxini-species groups of G. (Gyrodactylus) (Malmberg, 1970). Based on the morphology of the ventral bar and marginal hooks, G. markakulensis seems to fit into the G. phoxini-group. However, there are some specific characteristics that may support exclusion of this species from the G. phoxini-group or even the G. (Gyrodactylus) subgenus, such as the specific shape of the marginal hook tip. Moreover, there were differences in penis morphology; the penis of G. markakulensis is typified by rows of fine penis spines, finer than those of other Gyrodactylus subgenera. However, this is a plesiomorphic character shared with G. sedelnikowi and the other species of the G. (Gyrodactylus) subgenus, typified by a row of these fine spines and a row of larger penis spines.
The monophyletic origin of the other 5 Gyrodactylus subgenera, based on the present analyses, is also controversial. They all fall into one well-supported clade as a sister group to G. (Gyrodactylus) and G. markakulensis. Within this clade, 2 groups are recognized, separating species of G. (Mesonephrotus) and G. (Metanephrotus) from those of G. (Paranephrotus), G. (Neonephrotus) and G. (Limnonephrotus). None of these subgenera were found to be monophyletic. Within the G. (Metanephrotus) and G. (Mesonephrotus) clade, there are 2 well-supported associations; the first consists of Gyrodactylus turnbulli, Gyrodactylus poeciliae and Gyrodactylus bullatarudis. These species are specific parasites of fish from the genus Poecilia and their close relationship was previously suggested by Harris & Cable (2000). The excretory system of G. turnbulli was described as a G. (Metanephrotus)-like system (Harris, 1986), and, based on the morphology of the marginal hooks and ventral bar, this species falls into the G. eucaliae-group (Cable et al. 1999). However, based on the definition of G. (Metanephrotus) (Malmberg, 1970), there is no strong evidences of convincing autapomorphies to place this subgenus as ‘more derived’ as presented by Malmberg (1998). Nevertheless, the position of species within this clade might be biased by the fact that only a few species of the G. eucaliae-group were sequenced, and also by different mechanisms such as host–parasite coevolution that may play an important role.
The third well-supported clade consists of species of the G. (Limnonephrotus), G. (Paranephrotus) and G. (Neonephrotus) subgenera. Species of the 2 latter subgenera clustered together in a terminal position of the tree and this could be a consequence of the limited number of species sequenced. The morphology of the ventral bar and marginal hook of species of the G. (Paranephrotus) subgenus is similar to the majority of species of the G. (Limnonephrotus) subgenus. In addition, the marginal hook of G. anguillae is also of similar shape to G. (Paranephrotus) species but the ventral bar lacks lateral processes. Some of the terminal resolutions, especially the group of (Gyrodactylus vimbi – Gyrodactylus luciopercae) are also worth mentioning, as the morphology of the attachment apparatus is very similar and might support the idea of species groups based on the shape of the marginal hook and ventral bar. Close relationships among the majority of species in the group have been discussed already, and the G. wageneri-species group to which these species belong was considered as monophyletic, as was the G. (Limnonephrotus) subgenus (Zietara & Lumme, 2002). A greater number of species of the G. wageneri-group were included in the present study (especially species parasitizing cyprinids), and the monophyletic origin of the G. wageneri-group and the G. (Limnonephrotus) subgenus was rejected.
Zietara et al. (2002) claimed that analysis of ITS sequence revealed deep divisions within the genus Gyrodactylus that followed Malmberg's (1970) phylogeny. This study has shown that the monophyly of groups demonstrated by Zietara et al. (2002) cannot be supported, and may have been a result of the low number of species studied. Their conclusions, based on analysis of only 10 from a genus that contains over 400 species, appear to have been premature, and the close grouping of the G. wageneri-group species found by Zietara & Lumme (2002) may be expected from the species studied, which represented restricted host and geographical ranges. Future studies may reveal similar deep divisions within this and other genera and it is likely that analysis of additional species of Gyrodactylus will produce more species groups that are difficult to resolve by use of ITS alone.
Finally, we conclude from the results of the present phylogenetic analyses that the characteristics of the excretory system of Gyrodactylus as presented by Malmberg (1970) do not seem to be sufficiently conservative or informative to reveal subgenera within Gyrodactylus. Moreover, it is impossible to use these characters when the excretory system is unknown in the majority of newly described species. The validity of species groups within this genus is supported, as the morphology of the ventral bar and marginal hooks seem to have power to inform us about relationships between Gyrodactylus species. However, we found that some authors do not place species in any species group as part of the species description and that comprehensive revisions might be necessary. The present phylogenetic analyses inferred from the complete ITS region give us satisfactory, although limited, resolution and different topologies may form within the terminal groups when other regions of DNA are analysed.
We would like to thank Marketa Ondrackova and Radim Blazek (Masaryk University, Czech Republic) for help with collecting material and Sandy Mitchell (FRS Marine Laboratory, Scotland) for kindly operating the automated sequencer. We would also like to thank Eva Rehulkova for providing material of Macrogyrodactylus polypterii. I. M. was supported by EC Marie Curie Individual Fellowship QLK5-CT-2001-51038, M. G. was supported by The Research Project of Masaryk University No. MSM 143-1000-10 and D. T. J. L. was supported by a Wellcome Trust Fellowship (043965).
References
REFERENCES

Table 1. Monopisthocotylean parasite sequences used for ML analysis of the SSU rRNA gene for outgroup comparison (Olson & Littlewood, 2002)

Table 2. Species used for phylogenetic analyses of the ITS

Fig. 1. Phylogeny of monopisthocotylean Monogenea based on SSU rDNA indicating the relative position of Gyrodactylus species and potential outgroup taxa. The tree topology is from a maximum likelihood analysis with nodal support indicated, from top to bottom, for maximum likelihood (bootstrap %, n=100), maximum parsimony (bootstrap %, n=1000) and Bayesian inference (posterior probabilities); see text for further details.

Fig. 2. Phylogeny of Gyrodactylus species based on ITS rDNA, rooted against Gyrodactyloides bychowskii. The tree topology is from a Bayesian analysis, modelling each data partition separately, with nodal support indicated, from top to bottom, for Bayesian inference (posterior probabilities) and maximum parsimony (bootstrap %, n=1000) where these values are >80% and 50% respectively. Subgenera, where known, are indicated in square brackets as: G. Gyro – G. Gyrodactylus; G. Limno – G. Limnonephrotus; G. Meso – G. Mesonephrotus; G. Meta – G. Metanephrotus; G. Neo – G. Neonephrotus; G. Para – G. Paranephrotus according to terms proposed by Malmberg (1956, 1970); see text for further details.
- 48
- Cited by


