Introduction
Members of the family Diplostomidae Poirier 1886, are endoparasites with a worldwide distribution. Recent morphological and molecular studies have contributed to our understanding of the diversity, evolution and host–parasite interactions of this enigmatic group of diplostomatids (see references in Blasco-Costa & Locke, Reference Blasco-Costa and Locke2017). However, these studies were focused mainly in the Palaearctic region, leaving gaps in the knowledge of this group of parasites in other biogeographical regions, such as in the Neotropics.
The genus Hysteromorpha Lutz, 1931 currently contains two species, the type species Hysteromorpha triloba (Rudolphi 1819) Lutz, 1931 and H. plataleae Dubinin et Dubinina, 1940 from Russia (see Dubois, Reference Dubois1970). As in other diplostomids, the species H. triloba parasitizes the intestines of fish-eating birds of the genera Phalacrocorax Brisson, Ardea L. and Nyctanassa L. across the globe (Hugghins, Reference Hugghins1954; Dubois, Reference Dubois1970). In the Americas, metacercariae have been reported primarily in cyprinid fishes, but they have also been reported in four other fish families: Ictaluridae, Catostomidae, Ariidae and Pimelodidae (see Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García Prieto and Mendoza Garfías2007; Drago et al., Reference Drago, Lunaschi and Schenone2011; Locke et al., Reference Locke, McLaughlin, Lapierre, Johnson and Marcogliese2011; Monteiro et al., Reference Monteiro, Amato and Amato2011), and the planorbid snail Gyralus hirsutus Gould has been reported as the first intermediate host of H. triloba (Hugghins, Reference Hugghins1954).
In Mexico, in the Neotropical region, adults and metacercariae of H. triloba were recorded at two localities from Pacific Ocean slopes. However, vouchers of those specimens were not deposited in any collection and the report could not be verified (see Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García Prieto and Mendoza Garfías2007). In the current study, specimen adults and metacercariae identified as H. triloba from the Neotropical cormorant (Nannopterum brasilianus Gmelin) and Mexican tetra fish (Astyanax mexicanus De Filippi) were collected from the Gulf of Mexico and Pacific Ocean slopes. The aims of this study were: (1) to characterize molecularly the adults and metacercariae of H. triloba; (2) to link the adult and metacercariae using sequences of both internal transcribed spacers plus 5.8S from nuclear ribosomal DNA, and cytochrome c oxidase subunit 1 from mitochondrial DNA; (3) to examine the ultrastructure of the body surface of adults and metacercariae using scanning electron microscopy; and (4) to provide a morphological description of both stages.
Materials and methods
Specimen collection
A total of 61 birds, including 47 Neotropical cormorants (N. brasilianus), and 14 double-crested cormorants (Nannopterum auritus Lesson) were collected between June 2005 and March 2015 in 25 localities across Mexico. However, in only two localities, one in the Gulf of Mexico (Tlacotalpan, Veracruz, 18°36′0″N, 95°39′0″W) and the other from Pacific Ocean slopes (La Angostura, Chiapas, 16°11′31″N, 92°59′52″W), two Neotropical cormorants were infected with five and 15 mature adults of H. triloba respectively. The intestines were placed in separate Petri dishes with 0.75% saline solution and examined under a dissecting microscope. Avian definitive hosts were identified using the field guides of Howell & Webb (Reference Howell and Webb1995) and the American Ornithologists’ Union (1998). Freshwater fishes from eight families (Atherinopsidae, Cichlidae, Eleotridae, Gobiidae, Heptapteridae, Mugilidae, Godeidae and Poeciliidae) were examined in search of metacercariae of H. triloba. However, only the Mexican tetra fish (A. mexicanus) from San Luis Potosi (100°17′24″N, 99°43′12″W), at a tributary from the Panuco River in the Gulf of Mexico, was positive for the infection. Fish were captured by electrofishing, maintained alive and transported to the laboratory, pith sacrificed and examined immediately. Collected digeneans were fixed, by sudden immersion in hot (steaming) 4% formalin, for morphological comparisons. The fish were identified following Miller et al. (Reference Miller, Minckley and Norris2005).
Morphological study
Unflattened digeneans preserved in formalin were stained with Mayer's paracarmine, dehydrated in a graded ethanol series, cleared with methyl salicylate and mounted as permanent slides in Canada balsam. Drawings were made with the aid of a drawing tube. Measurements are given in micrometres (μm) followed by the range.
The following measurements were taken from the specimens studied: body total length, forebody length, forebody width, hindbody length, hindbody width, oral sucker length, oral sucker width, ventral sucker length, ventral sucker width, left pseudosucker length, left pseudosucker width, right pseudosucker length, right pseudosucker width, pharynx length, pharynx width, oesophagus length, oesophagus width, holdfast organ length, holdfast organ width, proteolytic gland length, proteolytic gland width, ovary length, ovary width, anterior testis length, anterior testis width, posterior testis length, posterior testis width, egg length and egg width.
Specimen (paragenophore sensu Pleijel et al., Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) adults and metacercariae were deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México (UNAM), Ciudad de México, México, under numbers: 10631 for the adults and 10632 for the metacercariae. A subsample from these isolates was also fixed in 100% ethanol for molecular work.
Amplification, sequencing of DNA, alignments and phylogenetic analyses
A total of 15 specimens identified as H. triloba, 5 metacercariae and 10 adults (5 from the Gulf of Mexico and 5 from the Pacific Ocean slopes), were placed individually in tubes and digested overnight at 56°C in a solution containing 10 mm Tris–HCl (pH 7.6), 20 mm NaCl, 100 mm Na2 EDTA (pH 8.0), 1% Sarkosyl and 0.1 mg/ml proteinase K. Following digestion, DNA was extracted from the supernatant using the DNAzol reagent (Molecular Research Center, Cincinnati, Ohio, USA) according to the manufacturer's instructions. The cytochrome c oxidase subunit 1 (cox 1) of the mitochondrial DNA and the internal transcribed spacers (ITS1 and ITS2 plus 5.8S) from nuclear ribosomal DNA were amplified using the polymerase chain reaction (PCR). A fragment of cox 1 was amplified using the forward primer Plat-diploCOX1F, 5′-CGTTTRAATTATACGGATCC-3′ and the reverse primer Plat-diploCOX1R, 5′-AGCATAGTAATMGCAGCAGC-3′ (Moszczynska et al., Reference Moszczynska, Locke, McLaughlin, Marcogliese and Crease2009). The ITS region was amplified using the forward primer D1, 5′-GTCGTAACAAGGTTTCCGTA-3′ and the reverse primer D2, 5′-ATCTAGACCGGACTAGGCTGTG-3′ (Bowles & McManus, Reference Bowles and McManus1993). PCR reactions (25 μl) consisted of 2 μl of genomic DNA, 1 μl of each primer (10 pmol), 2.5 μl of 10× buffer, 1.5 μl 2 mm MgCl2, 0.5 μl of a mix of 10 mm deoxyribonucleoside triphosphates (dNTPs) and 1 U of Taq DNA polymerase (Platinum Taq, Invitrogen Corporation, São Paulo, Brazil). PCR cycling parameters consisted of denaturation at 94°C for 1 min; followed by 35 cycles of 94°C for 1 min, annealing at 50°C for 1 min and extension at 72°C for 1 min; followed by a post-amplification incubation at 72°C for 10 min. Sequencing reactions were performed using two initial and two internal primers for ITS, BD3 5′-GAACATCGACATCTTGAACG-3′ and BD4 5′-ATAAGCCGACCCTCGGC-3′, and two initial primers for cox 1 with ABI Big Dye (Applied Biosystems, Boston, Massachusetts, USA) terminator sequencing chemistry, and reaction products were separated and detected using an ABI 3730 capillary DNA sequencer. Contigs were assembled and base-calling differences resolved using Codoncode Aligner version 5.1.5 (Codoncode Corporation, Dedham, Massachusetts, USA). Sequences were deposited in the GenBank database under numbers MG649464–MG649478 for cox 1 and MG649479–MG649493 for ITS. Cox 1 and ITS sequences obtained in the current research were aligned with sequences available in GenBank of H. triloba and of other genera of diplostomids: Posthodiplostomum Dubois 1936, Mesoophorodiplostomum Dubois 1936, Ornithodiplostomum Dubois 1936, Diplostomum von Nordmann 1832, Austrodiplostomum Szidat & Nani, 1951 and Tylodelphys Diesing, 1850; in addition, sequences of the strigeids Cardiocephaloides Sudarikov 1959, Australapatemom burti Miller 1923, Parastrigea diovadena Dubois & Macko, 1972, Apharyngostrigea cornu Ciurea 1927 and Ichthyocotylurus, Odening 1969 were used as outgroups, because this family is considered as the sister group of Diplostomidae (see Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003; Hernández-Mena et al., Reference Hérnandez-Mena, García-Varela and Pérez Ponce de León2017). Sequences were aligned using Clustal W (Thompson et al., Reference Thompson, Gibson, Plewniak and Jeanmougin1997). Maximum likelihood (ML) and Bayesian inference analyses (BI) were performed for each dataset. The ML tree was inferred using RAxML 7.0.4. (Stamatakis, Reference Stamatakis2006). The best-fit nucleotide substitution models inferred with jModeltest (Posada, Reference Posada2008) were TIM3 + I + G for the cox 1 alignment and TVM + G for the ITS alignment. Tree searches were performed using 1000 (ML) random taxon addition heuristic searches. Clade support was assessed by bootstrap resampling with 10,000 replicates. Bayesian analyses were performed with MrBayes version 3.1.2 (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001). Settings were two simultaneous Markov chain Monte Carlo (MCMC) runs for 10 million generations, sampling every 1000 generations, a heating parameter value of 0.2 and a ‘burn-in’ of 25%. Trees were drawn using FigTree version 1.3.1 (Rambaut, Reference Rambaut2006). The genetic divergence among taxa was estimated using uncorrected ‘p’ distances with the program MEGA version 6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
Results
Morphological study of adults and metacercariae of Hysteromorpha triloba
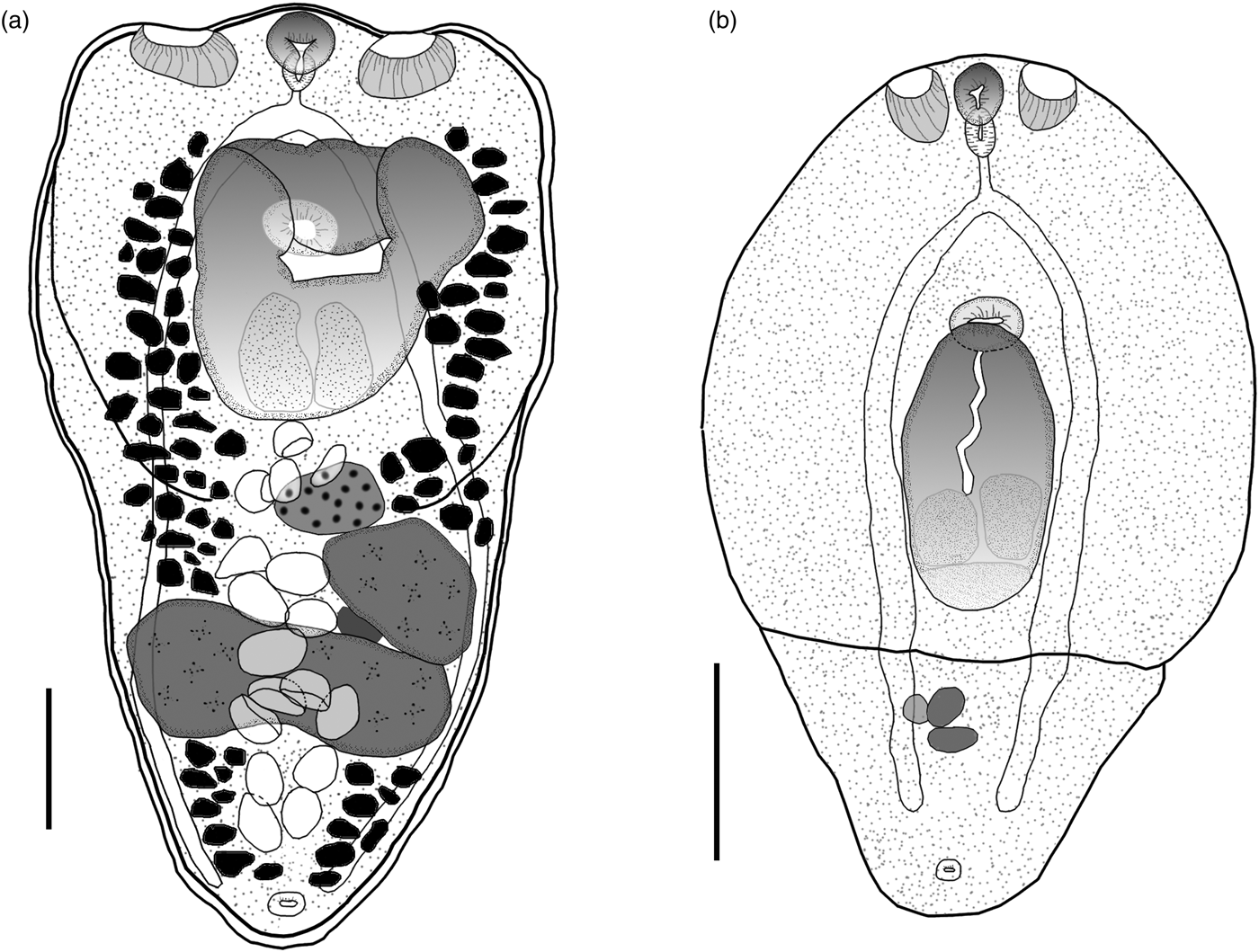
Eight adult worms identified as H. triloba obtained from N. brasilianus from La Angostura, Chiapas, were measured and morphologically characterized as follows. Body distinctly bipartite with spatulate shape. Total body length 1068–1333 (1220). Forebody trilobated, concave, 441–616 (558) long by 566–754 (665) wide (figs 1a, 2a). Hindbody subtriangular, 534–731 (657) long by 407–540 (465) wide. Oral sucker small, fairly muscular, subterminal, 72–88 (80) long by 77–95 (86) wide; two well-developed lateral pseudosuckers on each side of oral sucker. Left pseudosucker, 82–127 (105) long by 90–142 (120) wide. Right pseudosucker, 88–108 (98) long by 98–168 (128) wide. Ventral sucker oval, 80–100 (90) long by 90–109 (98) wide, situated immediately anterior to holdfast organ and sometimes covered by it (figs 1a, 2a). Prepharynx absent. Pharynx 47–70 (56) long by 40–51 (45) wide, elongate–oval, muscular. Intestinal bifurcation in anterior quarter of forebody. Caeca long, extending to the posterior end of hindbody. Holdfast organ elongate–oval, covered with numerous tiny spines (fig. 2b), 184–376 (286) long by 248–490 (337) wide. Proteolytic gland typically with bipartite appearance, situated at posterior margin of holdfast organ dorsally, 158–235 (207) long by 162–250 (183) wide. Testes in tandem, located in the anterior region of hindbody. Anterior testis ovoid, 130–194 (172) long by 134–227 (194) wide; posterior testis bilobulated, transversally elongated, 136–369 (201) long by 179–455 (366) wide. Ovary pretesticular, subspherical, 104–179 (140) long by 75–156 (105) wide, contiguous with anterior testis. Vitellarium in fore- and hindbody, beginning from anterior margin of holdfast. Vitelline reservoir intertesticular. Uterus distributed from posterior end of holdfast organ to anterior to the genital pore, which is subterminal. Eggs 77–98 (85) long by 46–63 (55) wide. Genital cone absent.
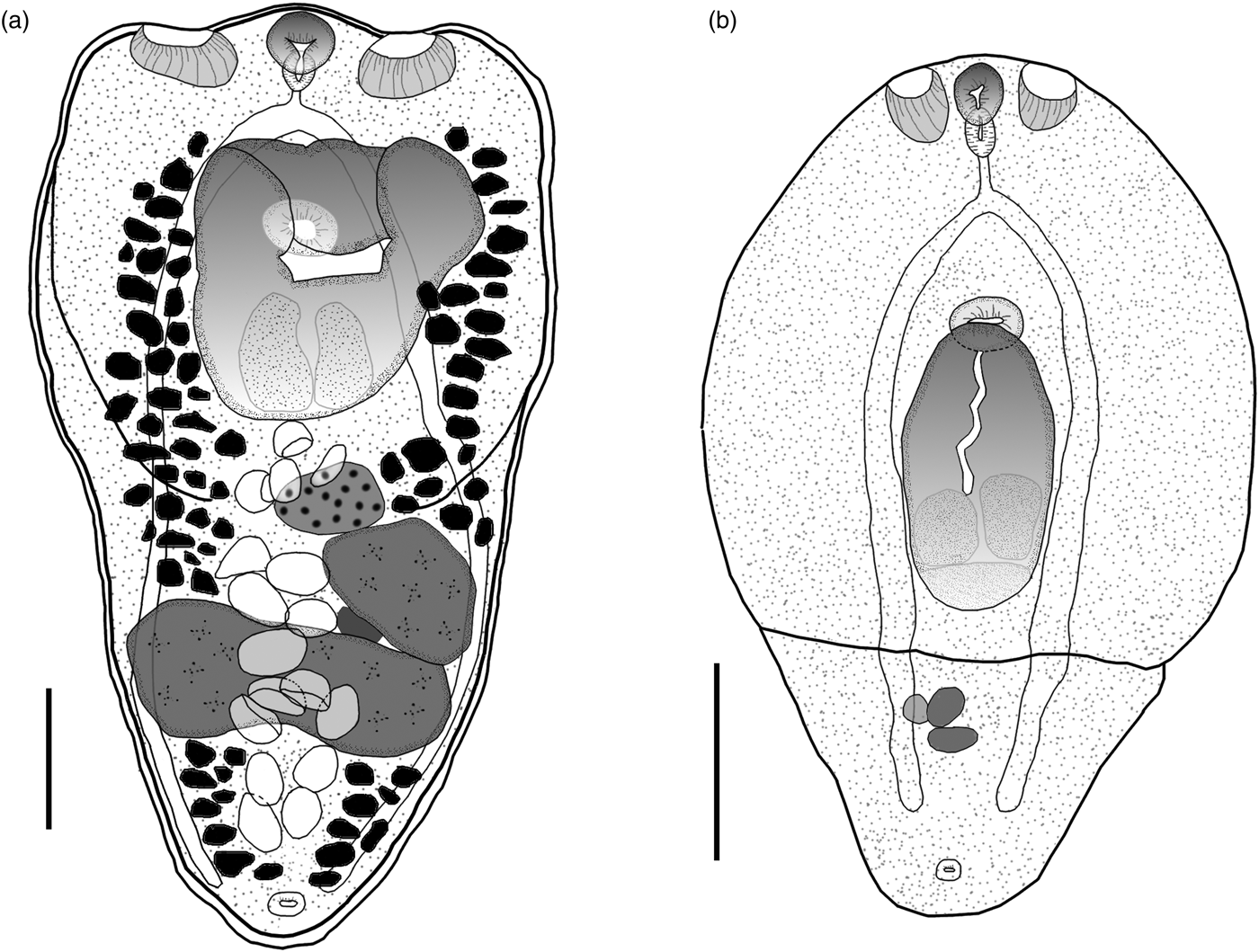
Fig. 1. Hysteromorpha triloba (Rudolpi, 1819), Lutz, 1931. (a) Adult obtained from the intestine of Nannopterum brasilianus. (b) Metacercaria obtained from the body cavity of Astyanax mexicanus. Scale bars: 200 μm.
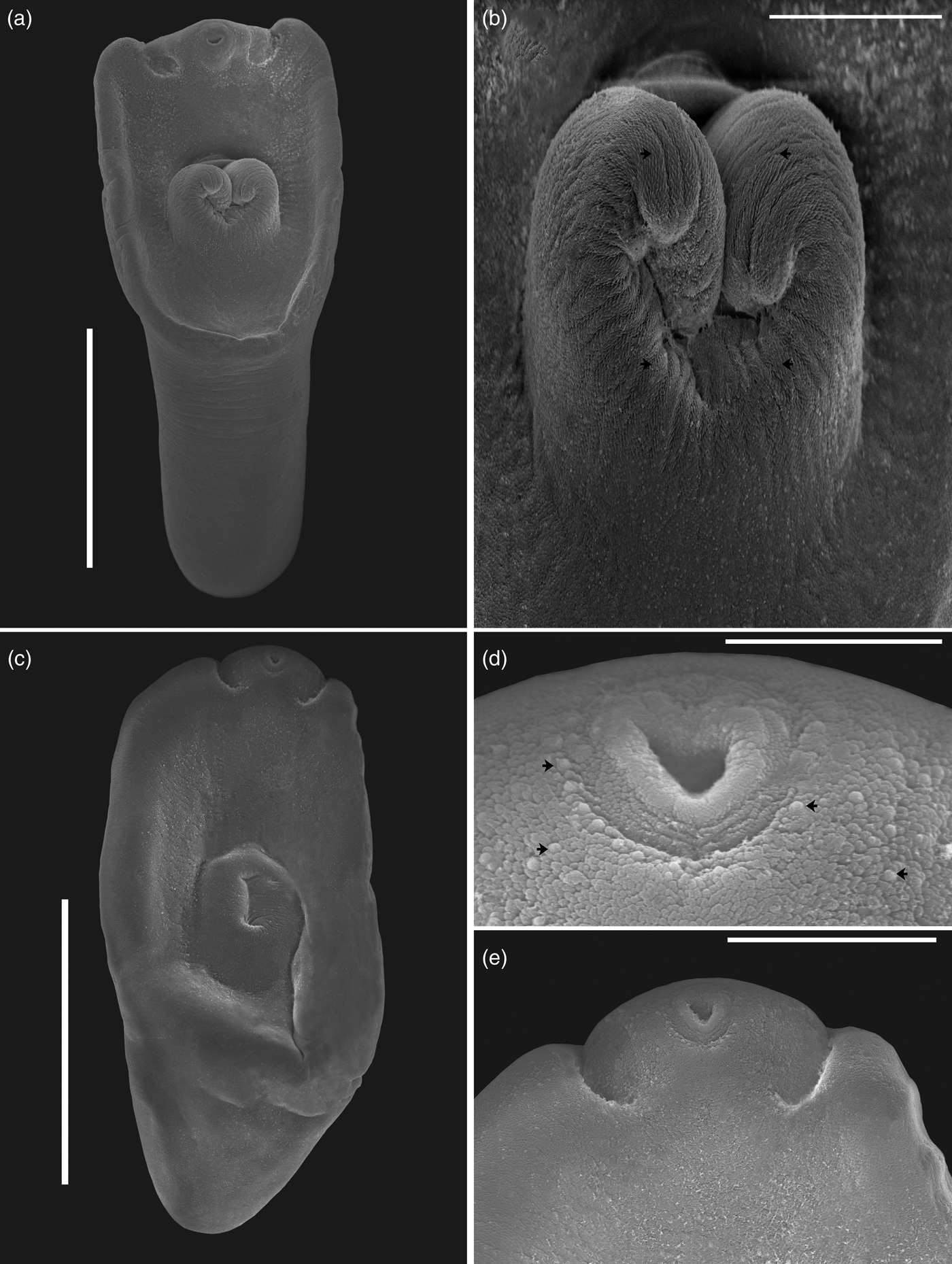
Fig. 2. Scanning electron micrographs of Hysteromorpha triloba (Rudolpi, 1819), Lutz, 1931. (a) Adult obtained from the intestine of Nannopterum brasilianus; (b) forebody region showing holdfast with spines, shown with black arrows. (c) Metacercaria from the body cavity of Astyanax mexicanus; (d) forebody showing the oral sucker recovered with papillae, shown with black arrows; (e) oral sucker and pseudosucker. Scale bars: (a) 400 μm; (b, e) 100 μm; (c) 300 μm; (d) 30 μm.
The following characterization is based on five metacercariae obtained from the Mexican tetra fish, A. mexicanus, from San Luis Potosi, Mexico. Body distinctly bipartite (figs 1b, 2c–e). Total body length 641–836 (732). Forebody elongate with ventral concavity, covered with numerous tiny papillae, 453–586 (547) long by 498–532 (517) wide (fig. 2d). Hindbody reduced to small conical prominence, 136–251 (188) long by 209–349 (289) wide. Oral sucker small, subterminal, 56–65 (61) long by 43–56 (52) wide. Two well-developed lateral pseudosuckers on each side of oral sucker (figs 1b, 2e). Left pseudosucker 59–90 (71) long by 45–60 (53) wide. Right pseudosucker 57–88 (70) long by 46–66 (55) wide. Ventral sucker oval, fairly muscular, 47–54 (50) long by 69–76 (72) wide; situated immediately anterior to holdfast organ. Prepharynx absent. Pharynx 42–53 (48) long by 24–35 (55) wide, elongate–oval, muscular. Intestinal bifurcation in anterior quarter of forebody. Caeca long, terminating at posterior level of primordial of testes. Holdfast organ 143–195 (179) long by 134–158 (146) wide, elongate–oval, opening longitudinally, lobulated, located near to posterior margin of forebody. Proteolytic gland typically with bipartite appearance, situated at posterior margin of holdfast organ dorsally, 48–71 (54) long by 106–136 (118) wide. Reproductive system poorly developed, with primordia of two oval testes. Anterior testis 27–74 (43) long by 36–75 (59) wide; posterior testis 22–38 (29) long by 32–67 (54) wide. Primordial ovary 29–48 (40) long by 69–86 (77) wide, placed at same level of anterior testis, elongate–oval. Genital pore subterminal, dorsal.
Remarks
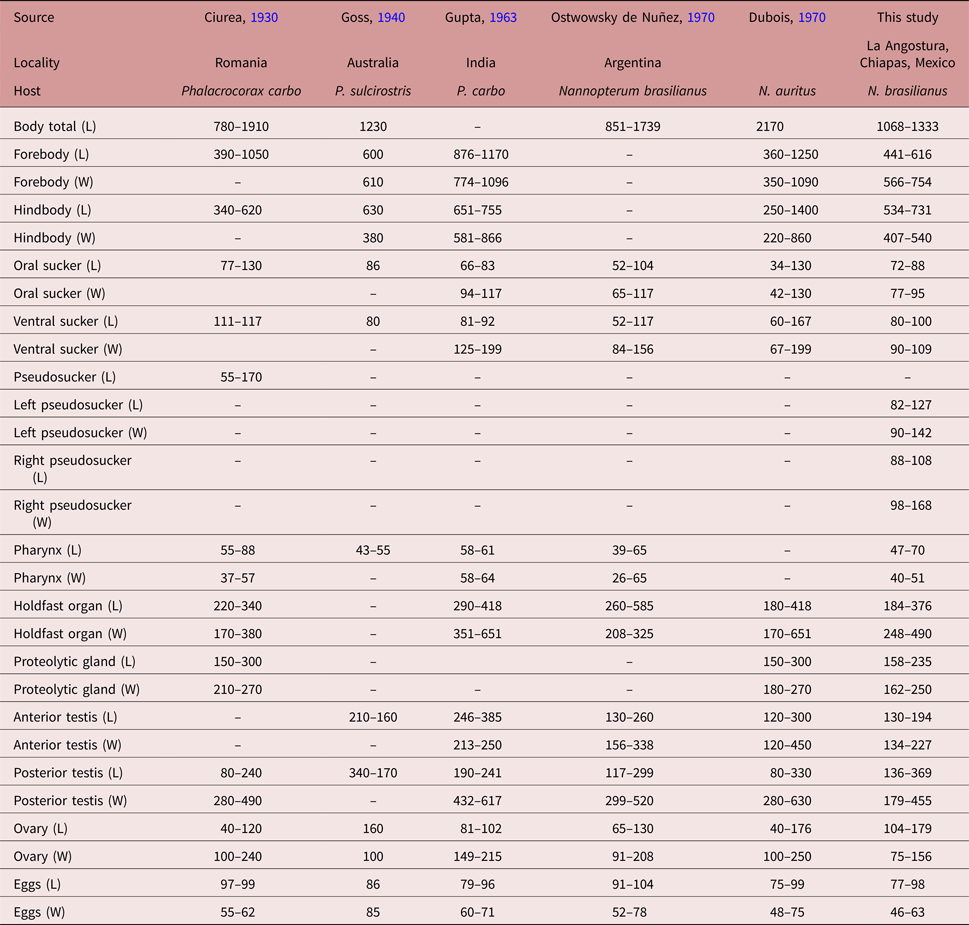
Following the original description of Lutz (Reference Lutz1931) and subsequent descriptions (Ciurea, Reference Ciurea1930; Goss, Reference Goss1940; Gupta, Reference Gupta1963; Ostrowski de Nuñez, Reference Ostrowski de Nuñez1970; Dubois, Reference Dubois1970), our specimens collected in two cormorants (N. brasilianus) from Mexico possess features that are consistent with the diagnosis of H. triloba: a distinctly bipartite body; forebody concave, trilobated; hindbody subtriangular; oral sucker with lateral lobes separated by two pseudosuckers; prepharynx absent; pharynx small; ventral sucker small; holdfast organ lobulated and covered with tiny spines; proteolytic gland bipartite; testes in tandem, anterior testis ovoid, smaller than posterior testis, which is bilobulated and transversally elongated; ovary pretesticular. However, our specimens show some level of morphological intraspecific variation. For instance, some meristic data of newly collected material had lower limits with respect to previous descriptions for the following characters: proteolytic gland width (162–250 vs. 180–270), anterior testis length (130–194 vs. 130–385), posterior testis width (179–455 vs. 280–630), ovary width (75–156 vs. 91–250) and egg width (46–63 vs. 48–78). Likewise, the newly collected material had higher limits for posterior testis length (136–369 vs. 80–299). In addition, we provided new measurements for each pseudosucker (see table 1).
Table 1. Comparative morphometrics (in microns) of adult worms of Hysteromorpha triloba (Rudolphi 1819) Lutz, 1931.
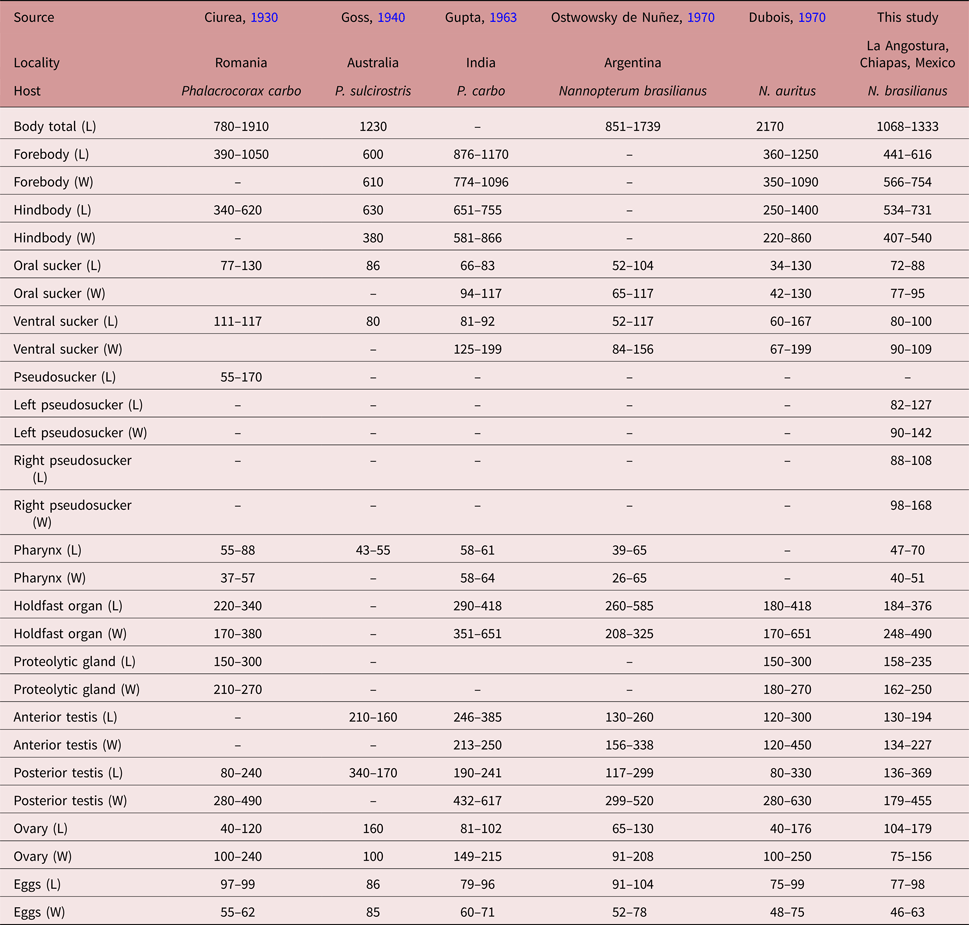
L, Length; W, width.
Molecular characterization
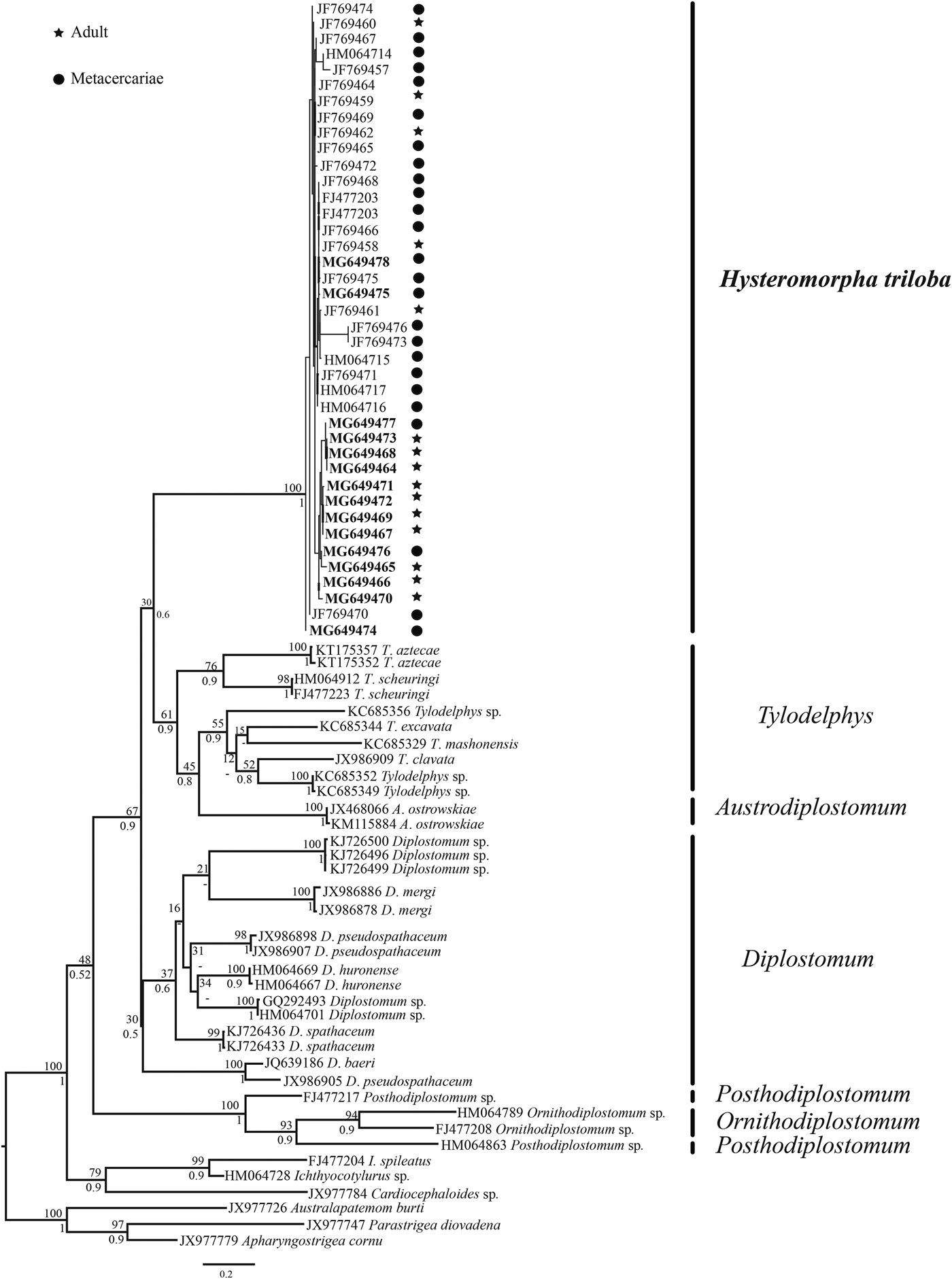
In this study, cox 1 sequences of 15 individuals of H. triloba (5 metacercariae and 10 adults) from the Neotropical region were generated and aligned with a cox 1 dataset that contained 24 isolates of H. triloba from the Nearctic region plus sequences of the following genera of diplostomids: Posthodiplostomum, Ornithodiplostomum, Diplostomum, Austrodiplostomum and Tylodelphys. The strigeids Cardiocephaloides, Australapatemon, Parastrigea, Apharyngostrigea and Ichthyocotylurus were used as outgroups. The alignment consisted of 97 sequences with 466 nucleotides. The genetic divergence among the genera of Diplostomatidae Posthodiplostomum, Ornithodiplostomum, Diplostomum, Tylodelphys and Austrodiplostomum ranged from 11 to 23%, and among species of the same genus ranged from 10 to 15%. The genetic divergence among 15 individuals of H. triloba (5 metacercariae and 10 adults) from the Neotropical region ranged from 0 to 2.5%, whereas the genetic divergence among the 24 isolates of H. triloba from the Nearctic region ranged from 0 to 4.7%. Finally, the genetic divergence among 39 individual of H. triloba ranged from 0 to 5.5%. Maximum likelihood (ML) analysis yielded a single tree that was identical in topology to the Bayesian inference (BI) consensus tree (fig. 3). The ML and Bayesian consensus trees showed that all the sequences of H. triloba generated in this study (10 adults and 5 metacercariae) are nested within a monophyletic clade, with strong bootstrap support and Bayesian posterior probability values (100/1.0). This clade also included the sequence of H. triloba from the double-crested cormorant from the Nearctic region (fig. 3).

Fig. 3. Maximum likelihood tree inferred with the cox 1 dataset; numbers near internal nodes show ML bootstrap clade frequencies and posterior probabilities (BI). The GenBank accession numbers in bold were generated in this study.
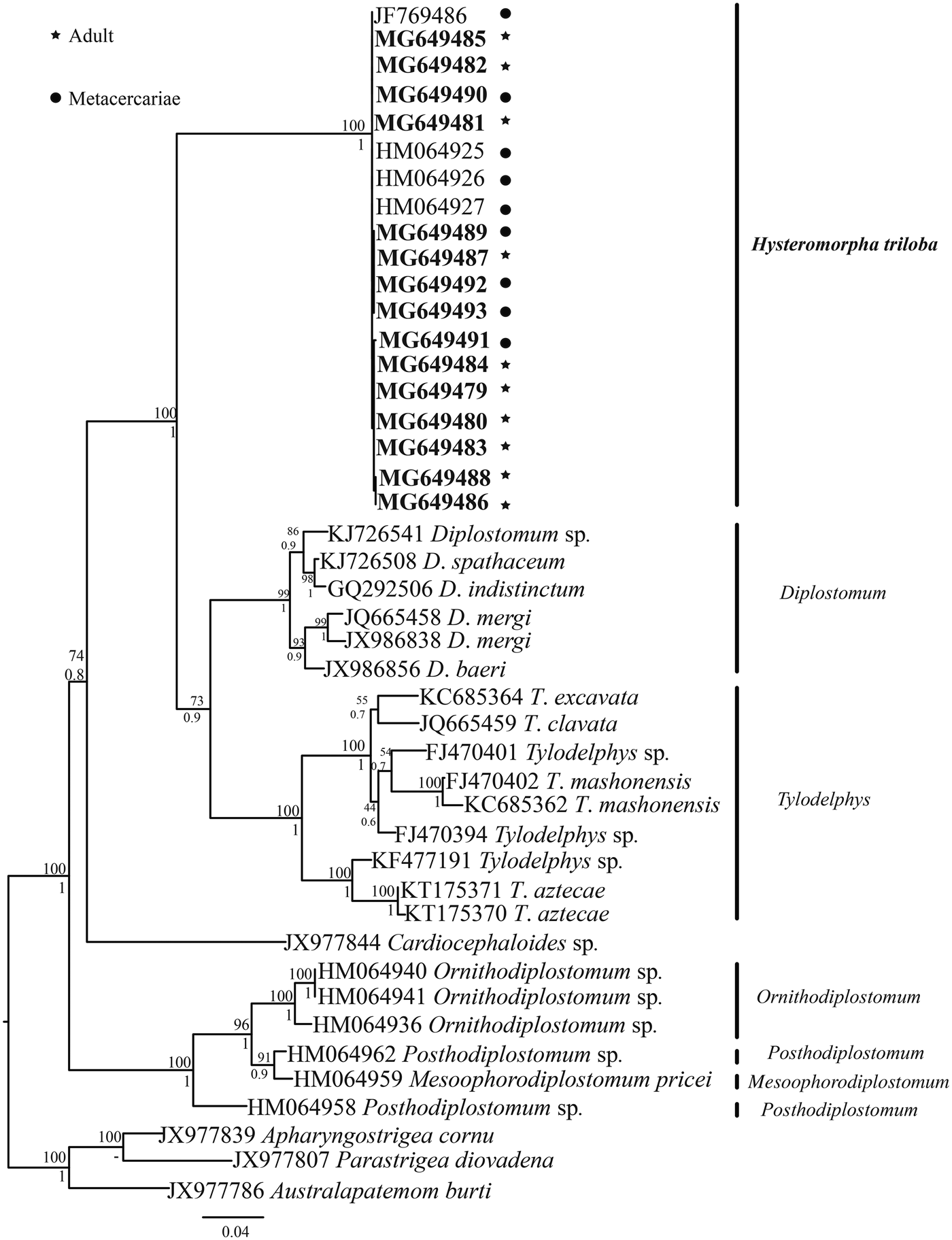
The ITS sequences of H. triloba (10 adults and 5 metacercariae) were aligned with four isolates of H. triloba from the Nearctic region and with sequences of other genera of Diplostomatidae. The alignment consisted of 44 sequences with 1138 nucleotides. The genetic divergence among the genera Posthodiplostomum, Ornithodiplostomum, Diplostomum, Hysteromorpha and Tylodelphys ranged from 14 to 18%, and among congeneric species of Diplostomum and Tylodelphys from 3 to 11%. In comparison, the genetic divergence among the 19 isolates of H. triloba was very low, from 0 to 0.2%. Maximum likelihood (ML) analysis yielded a single tree that was identical in topology to the Bayesian inference (BI) consensus tree (fig. 4). The ML and Bayesian consensus trees showed that all the sequences of H. triloba generated in this study (10 adults and 5 metacercariae) are nested within a clade, with strong bootstrap support and Bayesian posterior probability values (100/1.0). This clade also included four sequences (JF769486, HM064925–927) of H. triloba from the double-crested cormorant from the Nearctic region (fig. 4).

Fig. 4. Maximum likelihood tree inferred with the ITS1, 5.8S and ITS2 dataset; numbers near internal nodes show ML bootstrap clade frequencies and posterior probabilities (BI). The GenBank accession numbers in bold were generated in this study.
Discussion
The phylogenetic trees obtained with both molecular markers placed the metacercariae found in the body cavity from the Mexican tetra fish and the adults from the Neotropical cormorant in a single clade, confirming that both stages of the life cycle are conspecific. The genetic divergence estimated among 15 individuals of H. triloba from the Neotropical region (10 adults and 5 metacercariae) with the cox 1 dataset was very low, ranging from 0 to 2.5%, and among specimens of H. triloba from the Nearctic region ranging from 0 to 5.5%. These values of genetic divergence are higher than those found at the intraspecific level in other diplostomatid species. For instance, the genetic divergence among isolates of Tylodelphys sp., T. mashonense, T. excavata, T. aztecae and T. scheuringi ranged from 0 to 1.4% (Chibwana et al., Reference Chibwana, Blasco-Costa, Georgieva, Hosea, Nkwengulila, Scholz and Kostadinova2013, Reference Chibwana, Nkwengulila, Locke, McLughlin and Marcogliese2015; Otachi et al., Reference Otachi, Locke, Jirsa, Fellner-Frank and Marcogliese2015; García-Varela et al., Reference García-Varela, Sereno-Uribe, Pinacho-Pinacho, Hernández-Cruz and Pérez-Ponce de León2016; Blasco-Costa et al., Reference Blasco-Costa, Poulin and Presswell2017), and among isolates of Diplostomum mergi, D. pseudospathaceum and D. baeri from 0 to 1.01% (Georgieva et al., Reference Georgieva, Soldánová, Pérez-Del-Olmo, Dangel, Sitko, Sures and Kostadinova2013; Selback et al., Reference Selbach, Soldánová, Georgieva, Kostadinova and Sures2015). Finally, genetic divergence among isolates of Uvulifer spinatus ranged from 0 to 1.8% (López-Jimenez et al., Reference López-Jimenez, Pérez-Ponce de León and García-Varela2017). With respect to ITS1, 5.8S and ITS2, the genetic divergence estimated among the 19 isolates (5 metacercariae, 10 adults from the Neotropical region plus 4 isolates from the Nearctic region) of H. triloba was very low, ranging from 0 to 0.2%. This range of genetic divergence is similar to those described previously for congeneric diplostomatids. For example, genetic divergence ranged from 0 to 1.4% between Tylodelphys sp. and T. mashonense (see Chibwana et al., Reference Chibwana, Blasco-Costa, Georgieva, Hosea, Nkwengulila, Scholz and Kostadinova2013, Reference Chibwana, Nkwengulila, Locke, McLughlin and Marcogliese2015), from 0 to 0.03% among specimens of T. aztecae (García-Varela et al., Reference García-Varela, Sereno-Uribe, Pinacho-Pinacho, Hernández-Cruz and Pérez-Ponce de León2016), from 0.2 to 1.2% among isolates of Tylodelphys spp. (Blasco-Costa et al., Reference Blasco-Costa, Poulin and Presswell2017) and from 0 to 0.4% among isolates of D. baeri (see Blasco-Costa et al., Reference Blasco-Costa, Faltynková, Goergieva, Skirnisson, Scholz and Kostadinova2014). Finally, among isolates of Uvulifer spinatus genetic divergence ranged from 0 to 1.4% (López-Jimenez et al., Reference López-Jimenez, Pérez-Ponce de León and García-Varela2017).
Currently, Hysteromorpha is a genus distributed across the globe as parasite of fish-eating birds (see Hugghins, Reference Hugghins1954; Dubois, Reference Dubois1970; Ostrowski de Nuñez, Reference Ostrowski de Nuñez1970). The taxonomic history of the type species (H. triloba) has been unstable since its erection. For instance, it was described as Distomum trilobum, and was later transferred to the genera Hemistomum and Proalaria (see Dubois, Reference Dubois1970). Finally, Lutz (Reference Lutz1931) evaluated morphological characters based on its life cycle and transferred it to the new genus Hysteromorpha. Since then, H. triloba has been recorded in several countries, such as Argentina, Brazil, Venezuela, Mexico, USA and Canada in the Americas, associated with the Neotropical cormorant N. brasilianus and the double-crested cormorant N. auritus, suggesting that H. triloba can be regarded as a member of the ‘core’ helminth fauna of these two fish-eating bird species (Fedynich et al., Reference Fedynich, Pence and Bergan1997; Drago et al., Reference Drago, Lunaschi and Schenone2011; Locke et al., Reference Locke, McLaughlin, Lapierre, Johnson and Marcogliese2011; Monteiro et al., Reference Monteiro, Amato and Amato2011; O´Hear et al., Reference O'Hear, Pote, Yost, Doffitt, King and Panuska2014; Sheehan et al., Reference Sheehan, Hanson-Dorr, Dorr, Yarrow and Johnson2016). In contrast, the metacercaria of H. triloba exhibits low host specificity, since it has been recorded in at least eight species of freshwater fishes from unrelated families, such as Cyprinidae, Characidae, Catostomidae, Ictaluridae, Ariidae and Pimelodidae (see Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García Prieto and Mendoza Garfías2007; Locke et al., Reference Locke, McLaughlin, Lapierre, Johnson and Marcogliese2011).
Blasco-Costa & Locke (Reference Blasco-Costa and Locke2017) pointed out the great progress that has been made in recent years in the understanding of the diversity and evolution of diplostomatids, mainly in the Palaearctic region. This progress has left a big gap in the knowledge of this enigmatic group of parasites in the Neotropical region. Therefore, the current study contributes to our understanding of the genetic diversity, host–parasite interactions and life cycle of H. triloba in this biogeographical region.
Acknowledgements
We thank Berenit Mendoza Garfias for her help in obtaining the scanning electron microphotographs.
Financial support
This research was supported by grants from the Programa de Apoyo a Proyectos de Investigación e Inovación Tecnológica (PAPIIT-UNAM) IN206716. A.L.-J. and L.A.-G. thank the Programa de Posgrado en Ciencias Biológicas, Universidad Nacional Autonoma de México (UNAM) and Consejo Nacional de Ciencia y Tecnología (CONACYT) for the scholarships to complete their Master's degrees.
Conflict of interest
None.
Ethical standards
Specimens were collected in Mexico under the Cartilla Nacional de Colector Científico (FAUT 0202) issued by the Secretaría del Medio Ambiente y Recursos Naturales (SEMARNAT) to M.G.V.