Carbapenem-resistant organisms (CROs) are an important cause of healthcare-acquired infections and are particularly concerning because they are associated with high morbidity and mortality.Reference Amit, Mishali, Kotlovsky, Schwaber and Carmeli1–Reference Kumar, De, Baveja and Gore6 Carbapenem-resistant Enterobacteriaceae (CRE) have received significant attention,7 but glucose non-fermenting (NF) gram-negatives, such as Acinetobacter baumannii and Pseudomonas aeruginosa, are an additional and increasingly recognized carbapenem resistance reservoir.Reference Hong, Bae, Jang, Jeong, Kang and Lee8, Reference Kim, Kim, An, Cho, Park and Jang9 Of particular concern among CROs is the subset of carbapenemase-producing organisms (CPOs), for which carbapenem resistance is generally plasmid-mediated and which can transfer between organisms and across bacterial species. CPOs have been implicated in high-profile healthcare-associated outbreaksReference Snitkin, Zelazny and Thomas10 and may be associated with poorer clinical outcomes than non-CPOs.Reference Tamma, Goodman and Harris11
Admission screening for CRO and/or CPO carriage enables prompt implementation of isolation precautions for colonized patients and may provide an opportunity for individualized care, such as targeted empiric antibiotic therapy.Reference Dickstein, Edelman, Dror, Hussein, Bar-Lavie and Paul12–Reference Macesic, Gomez-Simmonds and Sullivan14 The Centers for Disease Control and Prevention (CDC) recommends CRE colonization screening in limited instances,15 but most US hospitals do not perform routine CRE or CRO screening. Given limited data on inpatient colonization prevalence in non-outbreak periods and current limitations of CRO and CPO diagnostics, universal screening remains impractical in many acute-care settings. However, recent CRE data indicate that existing targeted screening policies (eg, for recent foreign hospitalization, on direct transfer from outside facilities) miss many colonized patients.Reference Shimasaki, Segreti, Tomich, Kim, Hayden and Lin16, Reference Goodman, Simner and Klein17
Better identifying predictors of colonization and developing algorithms to predict colonization probability may improve targeted screening approaches. Existing strategies often rely on risk factors (ie, “independent” variables), but strong risk factors may not be good predictors. Our objectives were to measure the prevalence of CRO and CPO perirectal colonization at hospital unit admission and to develop machine learning–derived decision trees to predict patients’ probability of organism carriage.
Methods
Study setting and population
This study included patients aged ≥16 years admitted to the Johns Hopkins Hospital (JHH) medical intensive care unit (MICU) or solid organ transplant (SOT) unit between July 1, 2016 and July 1, 2017. Both units have longstanding vancomycin-resistant Enterococcus (VRE) surveillance programs and collect patient perirectal Eswabs (COPAN Diagnostics, Murrieta, CA) at unit admission (defined as ≤2 calendar days from unit entry) and weekly thereafter. This study was approved by the Johns Hopkins University School of Medicine Institutional Review Board, with a waiver of informed consent.
Microbiology methods and outcome definitions
Residual Amies media from Eswab collection vials was stored at 4°C, and within 4 days of swab collection, 100 µL was streaked for isolation onto a MacConkey agar with ertapenem and meropenem disks.Reference Simner, Martin, Opene, Tamma, Carroll and Milstone18 Colonies growing within 27 mm of ertapenem and within 32 mm of meropenem were identified by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Billerica, MA). Carbapenem antimicrobial susceptibility testing (ertapenem, meropenem, and imipenem) was performed by disk diffusion applying Clinical and Laboratory Standards Institute guidelines.19
Enterobacteriaceae resistant to ertapenem, meropenem, or imipenem were categorized as CRE. Glucose NF gram-negative bacilli resistant to meropenem and/or imipenem were categorized as NFCROs. Stenotrophomonas maltophilia was excluded due to its intrinsic carbapenem resistance. All CROs were tested for carbapenemase production using the modified carbapenem inactivation method (mCIM).Reference Pierce, Simner and Lonsway20 CRE and NFCROs positive for carbapenemase production by the mCIM test were defined as CP-CRE and CP-NFCROs, respectively (collectively called CPOs). mCIM-negative isolates were defined as non–CP-CRE and non-CP-NFCROs. CPOs underwent molecular carbapenemase genotype testing using the Check-MDR CT103XL assay (CheckPoints, Wageningen, Netherlands).
We coded all study laboratory data with a study identifier. We linked laboratory results and clinical data 6 months after sample collection or following patient discharge. Neither infection control nor clinical staff were aware of patient colonization status during the hospital admission.
Clinical data collection
Patient data were retrospectively collected using bulk extraction methods from the JHH electronic medical records (EMR) system and infection control and administrative databases. EMR data were available for inpatient and outpatient encounters across 5 Johns Hopkins Health System hospitals across Maryland and the District of Columbia. Extracted patient data included >125 variables capturing demographic, pre-existing medical condition, procedure, medication, and other clinical data (Table 1 and Supplemental Material online).
Table 1. Characteristics of Patients in the Medical Intensive Care Unit (MICU) and Solid Organ Transplant (SOT) Unit with Carbapenem-Resistant Organism (CRO) and Carbapenemase-Producing Organism (CPO) Perirectal Colonization at Unit Admission

Note. SD, standard deviation; ER, emergency room; IQR, interquartile range; NA, not applicable due to model nonconvergence or nonestimability. CRE, carbapenem-resistant Enterobacteriaceae; NFCRO, carbapenem-resistant glucose non-fermenting bacilli; HIV, human immunodeficiency virus; MDR, multidrug resistant; MDRO, multidrug-resistant organism; ESBL, extended-spectrum β-lactamase; VRE, vancomycin-resistant Enterococcus spp; MRSA, methicillin-resistant Staphylococcus aureus; WBC, white blood cell count.
a Table 1 does not include all variables and permutations evaluated in prediction models.
b Receipt of chemotherapy or immunosuppressive therapy in the prior 3 mo, HIV positive, and/or documented CBC immunosuppressive abnormalities within 24 h preceding unit admission (defined as absolute neutrophil counts or total WBC < 500 cells/mm3).
c Defined in reference to the National Healthcare Safety Network (NHSN) 2018 definition of “central line,” available at: https://www.cdc.gov/nhsn/pdfs/pscmanual/pcsmanual_current.pdf.
d Indications for contact precautions are a flagged history of: (1) MRSA; (2) VRE; (3) Clostridioides difficile; (4) MDR gram-negative bacteria; (5) CRE (which are classified separately from other MDR gram-negative bacteria at JHH); (6) respiratory viruses; and (7) other indications, including “CRE rule-out” for patients recently hospitalized internationally (≤6 mo prior to admission), enteric pathogens, and contact precautions without associated infection control flag(s).
e Resistant to 4 of 5 antibiotic classes tested.
f Immunosuppressant or nontopical glucocorticoid.
g Proton-pump inhibitors (PPIs) or histamine H2-receptor antagonists (H2-blockers). These medications were analyzed as a composite category in logistic regression, but were evaluated both individually and as a composite category in predictive models.
* Significant at P ≤ .05; **significant at P ≤ 0.01; ***significant at P ≤ .001, based upon a 2-tailed significance test in univariable logistic regression with general estimating equations and robust standard errors to account for patient-clustering due to repeat unit admissions.
Statistical methods
Data analysis and logistic regression
Descriptive statistics for patient variables were calculated using mean (standard deviation [SD]), median (interquartile range [IQR]), or frequency count (percentage), as appropriate, with Clopper-Pearson binominal 95% confidence intervals (CIs) for proportions. We compared CRO colonization at admission among MICU patients and SOT unit patients using the Fisher exact test. The relationship between each covariate and the study outcomes was evaluated using univariable logistic regression with general estimating equations and robust standard errors to account for patient clustering due to repeat unit admissions. Descriptive and logistic regression analyses were performed in Stata version 13.0 software (StataCorp, College Station, TX).
Machine learning-derived prediction models and validation
Using all collected variables (n = 134), we developed prediction models for the outcomes of CRO, CRE, and CPO colonization at unit admission. We built decision trees applying the classification and regression tree (CART) algorithmReference Breiman, Friedman, Stone and Olshen21 using the rpart (recursive partitioning and regression trees) package, version 4.1–13. To fit our trees, we employed the Gini impurity criterion for splitting rules.Reference Duda, Hart and Stork22 Ensemble-based decision tree learning methods were utilized in sensitivity analyses (see Supplemental Material online). All machine learning models were developed using the R statistical package version 3.0.5 (R Foundation for Statistical Computing, Vienna, Austria). CART decision trees were internally evaluated using leave-one-out cross-validation.Reference Duda, Hart and Stork22 The discrimination of all models, both original (in sample) and cross-validated (out of sample), were assessed through the generation of receiver operating characteristic (ROC) curves and the calculation of C statistics in R software.
Results
Study population
This study included 3,327 unit admissions during the one-year study period: 1,796 (54%) in the MICU and 1,531 (46%) in the SOT unit. Of these encounters, 2,878 (87%), representing 2,165 unique patients, had stored perirectal admission screening swabs that were processed for CROs (Fig. 1).
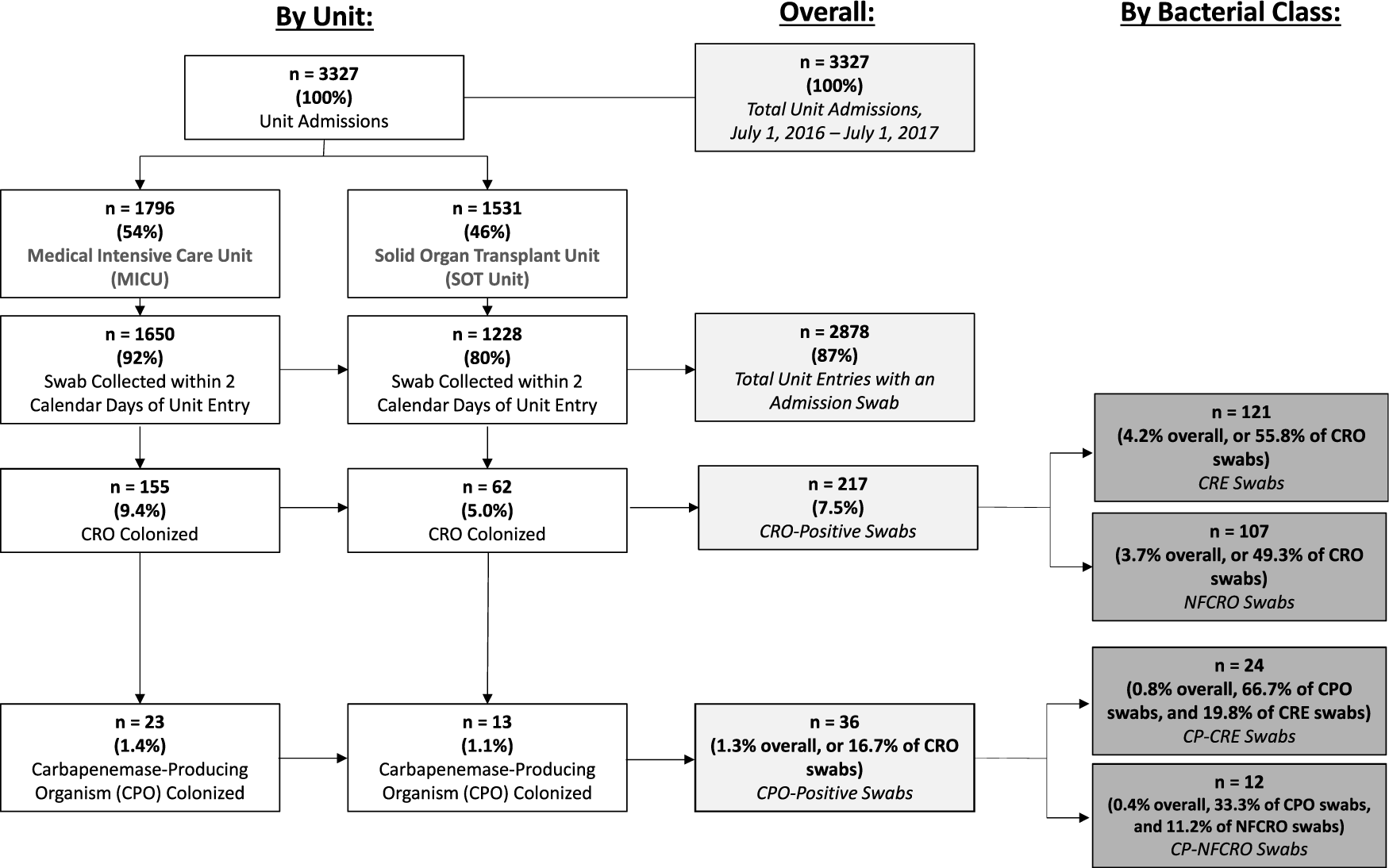
Fig. 1. Study flowchart of carbapenem-resistant organism (CRO) and carbapenemase-producing organism (CPO) colonization at hospital unit admission. Note. CRE, carbapenem-resistant Enterobacteriaceae; NFCRO, non-fermenter carbapenem-resistant organism; CP-CRE, carbapenemase-producing CRE; CP-NFCRO, carbapenemase-producing NFCRO.
Patient characteristics are presented in Table 1. In the 6 months preceding unit admission, 54% of patients had been hospitalized, 17.5% had a prior ICU stay, 6.0% had been discharged to a postacute care facility, and 1.0% of patients had documented overnight hospitalization in a foreign country. In the prior 3 months, 21.1% of patients had received antibiotics with gram-negative coverage, including 4.5% with carbapenems.
CRO and CPO colonization admission prevalence
Overall, 217 swabs (7.5%; 95% CI, 6.6%–8.5%) from 192 unique patients tested positive for 1 or more CROs (Fig. 1). Prevalence was higher among MICU admissions (9.4%; 95% CI, 8.0%–10.9%) than among SOT unit admissions (5.0%; 95% CI, 3.9%–6.4%; P < .001). Of the CRO-positive swabs, 36 (16.7%) from 32 unique patients demonstrated carbapenemase production, yielding a CPO colonization admission prevalence of 1.3% (95% CI, 0.9%–1.7%).
In total, 121 admission swabs from 113 unique patients were positive for CREs. The overall prevalences of CRE and CP-CRE perirectal colonization at admission were 4.2% (95% CI, 3.5%–5.0%) and 0.8% (95% CI, 0.5%–1.2%), respectively (Fig. 1). Twenty percent of CRE isolates were carbapenemase-producers.
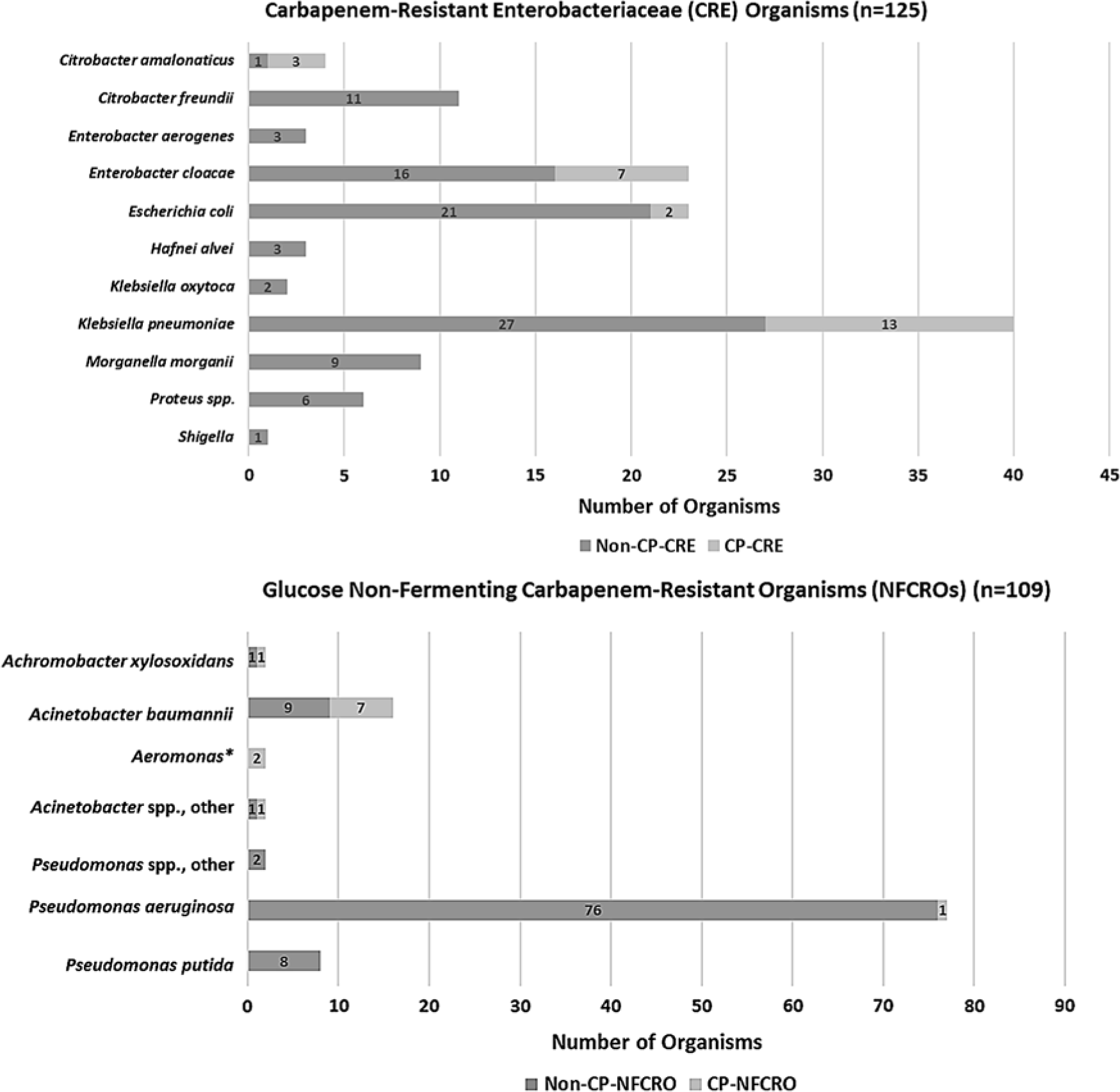
One hundred and seven admission swabs from 92 unique patients tested positive for 1 or more NFCROs. The overall prevalences of NFCRO and CP-NFCRO perirectal colonization at admission were 3.7% (95% CI, 3.0%–4.4%) and 0.4% (95% CI, 0.2%–0.7%), respectively. Eleven percent of NFCRO isolates were carbapenemase-producers. The distribution of CRE and NFCRO organisms by bacterial class and carbapenemase production status is provided in Fig. 2.
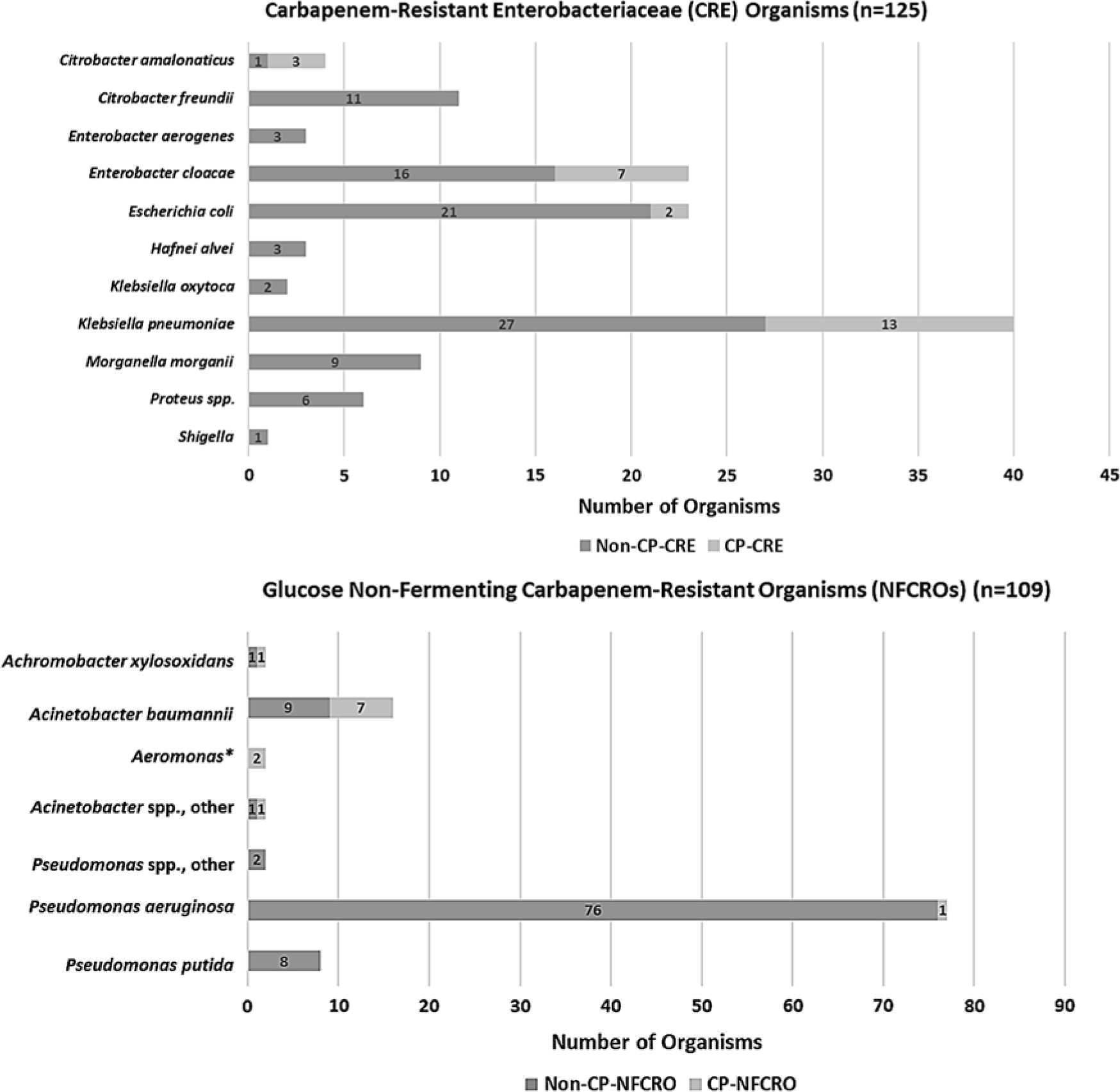
Fig. 2. Distribution of organisms by bacterial class and carbapenemase-production status (defined by a positive mCIM test), among 217 perirectal unit admission swabs positive for carbapenem-resistant organism (CRO) colonization. Overall, 20% of carbapenem-resistant Enterobacteriaceae (CRE) organisms, accounting for 24 swabs from 22 unique patients, were carbapenemase-producers (CP-CREs). Of 109 NFCROs, 12 (11.0%), accounting for 12 swabs from 10 unique patients, were carbapenemase-producers (CP-NFCROs). In addition, 11 admission swabs (0.4%), all from unique patients, were co-colonized with CRE(s) and NFCRO(s); 3 of these swabs possessed a carbapenemase-producing organism (CPO), but no admission swabs were CP-CRE and CP-NFCRO co-colonized. *Aeromonas categorized with glucose non-fermenting gram-negative bacilli for purposes of this study.
Overall, 33 organisms from 32 of 36 CPO-positive swabs (1 swab was co-colonized with 2 CP-CREs), as defined by a positive mCIM test, underwent molecular genotyping. Of the 33 processed mCIM-positive organisms, 23 (70%) were positive for carbapenemase genes by the Check-MDR CT103XL assay. The gene distribution was as follows: KPC (12, 52%), NDM (2, 9%), VIM (1, 4%), OXA-23 (2, 9%), OXA-24 (2, 9%), OXA-48-like (1, 4%), and NDM + OXA-48-like (3, 13%) (Supplemental Table 1 online).
Characteristics of patients with CRO and CPO colonization at unit admission
A large proportion of exposures were associated with CRO colonization (Table 1), including ostomy within 3 months of unit admission, history of a CRE-positive or NFCRO-positive culture in the prior 6 months, carbapenem or gastric acid suppressant (proton-pump inhibitor (PPI) or histamine H2-receptor antagonist) use in the prior 3 months, and post-acute care facility exposure (direct-admission from a skilled nursing/rehabilitation facility, or discharge to a long-term acute-care hospital or skilled nursing/rehabilitation facility in the prior 6 months).
When we restricted our analysis to the subset of CPO-colonized patients, the preceding variables remained associated with CPO colonization (Table 1). Additional variables were also associated with CPO colonization, including colorectal surgery in the prior 3 months and foreign travel by the patient or a partner in the preceding 21 days. Nevertheless, only 2 patients with molecularly confirmed carbapenemases (a KPC-producing K. pneumoniae and an OXA-48-like–producing K. pneumoniae) had documented foreign travel; none reported recent international hospitalization.
Both CRO- and CPO-colonized patients were significantly more likely than non-carriers to be on contact precautions at unit admission; however, only 46.1% of CRO carriers were on contact precautions at unit admission. CRO and CPO carriers were also more likely to test positive for VRE colonization during admission screening, that is, on the same swab that underwent CRO processing. These VRE-colonized patients accounted for 24% and 33% of CRO and CPO colonizations, respectively.
Predicting probability of colonization at unit admission
We evaluated all collected study variables, including permutations (eg, varied retrospective periods and composite and individual variable categories) for inclusion in decision tree models. These machine learning approaches are well suited to large EMR datasets because they can accommodate high predictor-to-outcome ratios, variable collinearities, and interaction effects by default.Reference Breiman, Friedman, Stone and Olshen21, Reference Roth, Battegay, Juchler, Vogt and Widmer23 Using branching logic rather than calculations, decision trees are also relatively user-friendly for manual bedside use. We derived models for 3 alternate outcomes: CRO, CPO, and CRE colonization (see Supplemental Material online for CRE colonization models).
The final decision tree for predicting CRO colonization at unit admission included 3 study variables (Fig. 4). The first question in the tree (“root node”), which is reserved for the most discriminatory variable, asked (1) “Did the patient have a CRO-positive culture in the previous 6 months?” If the answer was “yes,” the second question queried (2) “Did the patient receive 26 or more days (model-derived cut-point) of PPIs in the prior 3 months?” Patients meeting these criteria were classified as CRO positive with 93% probability (terminal node 4). In patients with a CRO history but lacking this PPI exposure, the tree questioned (3) “Has the patient been hospitalized for 51 or more days (model-derived cut-point) in the prior 6 months?” If the answer was “yes,” patients were classified as CRO positive (terminal node 3, 80% probability), and if the answer was “no,” patients were classified as CRO negative (terminal node 2, 74% probability).

Fig. 4. Decision tree for predicting CPO perirectal colonization at hospital unit admission. The gray-shaded terminal node indicates that the tree would classify patients as CPO colonized, and accompanying percentages reflect the probability that patients assigned to a given terminal node are CPO positive. Terminal node numbering, 1–2, is included in parentheses. The tree had an area-under-the-curve (C statistic) of 0.58, which was unchanged in cross-validation. Its sensitivity and specificity were 16.7% and 99.8%, respectively, and its positive and negative predictive values were 54.5% and 99.0%, respectively.
For the 2,804 patients lacking a recent CRO history, the root node branched left and terminated. Patients lacking this history were classified as CRO negative (terminal node 1, 93% probability).
The overall tree possessed a sensitivity of 9.8% and a specificity of 99.9%. The positive and negative predictive values were 87.5% and 93.1%, respectively. Incorporating outcome probabilities based on terminal node impurities, the C statistic for the final tree trained on the full dataset was 0.57 and remained unchanged following cross-validation.
The CPO decision tree truncated at a single variable, history of a CRE-positive culture in the prior 6 months (Fig. 3). Its sensitivity was 16.7% and its specificity was 99.8%. The CPO tree’s discrimination was 0.58 (unchanged following cross-validation).

Fig. 3. Decision tree for predicting CRO perirectal colonization at hospital unit admission. Gray-shaded terminal nodes indicate that the tree would classify patients as CRO colonized, and accompanying percentages reflect the probability that patients assigned to a given terminal node are CRO positive. Terminal node numbering, 1–4, is included in parentheses. The tree had an area-under-the-curve (C statistic) of 0.57, which was unchanged in cross-validation. Its sensitivity and specificity were 9.8% and 99.9%, respectively, and its positive and negative predictive values were 87.5% and 93.1%, respectively.
To optimize model performance and address possible outcome misclassification, we performed multiple sensitivity analyses: (1) we built prediction models for CRO and CPO colonization with random forests analysis; (2) we refit CART trees to increase sensitivity by imposing a greater “cost” for misclassifying colonized patients as negative; (3) we refit CART trees restricting to first, unique patient encounters (n = 2,165); and (4) we reperformed CART and random forests analyses after restricting the CPO outcome to isolates with molecularly confirmed carbapenemases. With more complicated models in sensitivity analyses 1 and 2, performance improved by ∼15%–20%; performance in analyses 3 and 4 was similar to the primary analyses. Results are provided and discussed in the Supplemental Material online.
Discussion
Identifying CRO- and CPO-colonized patients at hospital unit admission could facilitate timely infection control interventions, such as implementing prompt contact isolation precautions for colonized patients to limit healthcare-associated transmission. Evaluating patients admitted to MICU and SOT units, we found that 7.5% and 1.3% of patients were perirectally colonized with CROs and CPOs, respectively. Among CROs, the distribution of CRE versus NFCROs was roughly similar (54% vs 46%), with a CRE admission prevalence of 4.2%. This estimate is higher than the proportion of CRE (3.1%) among clinical isolates reported to the National Healthcare Safety Network in 2015Reference Woodworth, Walters and Weiner24 and considerably higher than the 0.5% CRE admission prevalence recently reported at a Chicago tertiary-care hospital (2013 data from ICU populations).Reference Shimasaki, Segreti, Tomich, Kim, Hayden and Lin16 Importantly, most colonized patients (54%) were not on contact precautions at unit admission (for any indication), posing a potential reservoir for transmission during their unit encounter.
Our study included many variables known to be risk factors for CRO and CPO colonization or infection, including MDRO history,Reference Logan and Weinstein25–Reference Simner, Goodman, Carroll, Harris, Han and Tamma27 antibiotic exposure overall,Reference Bhargava, Hayakawa and Silverman28–Reference Predic, Delano, Tremblay, Iovine and Brown30 or to carbapenems specifically,Reference Torres-Gonzalez and Cervera-Hernandez31, Reference Armand-Lefevre, Angebault and Barbier32 post-acute care facility stay,Reference Cunha, Kassakian and Chan33, Reference Prabaker, Lin and McNally34 immunosuppression,Reference Bhargava, Hayakawa and Silverman28 endoscopy,Reference Predic, Delano, Tremblay, Iovine and Brown30, Reference Torres-Gonzalez and Cervera-Hernandez31, Reference Muscarella35 and indwelling hardware.Reference Bhargava, Hayakawa and Silverman28, Reference Cunha, Kassakian and Chan33, Reference Prasad, Labaze and Kopacz36 Despite including these known risk factors and >100 other variables, our constructed models did not highly accurately predict CRO and CPO colonization, with C statistics of 0.57 and 0.58, respectively. Performance improved by ∼15%–20% in sensitivity analyses, with more complicated models that may be less likely to replicate in other settings and which would be less practical as bedside tools. Despite suboptimal global performance, however, the CRO decision tree did, with high accuracy, identify certain higher-colonization risk patient populations: patients with recent CRO-positive cultures (≤6 months) who had either ≥26 days of PPI usage in the prior 3 months (93% colonization probability) or ≥51 days of hospitalization in the prior 6 months (80% colonization probability). This observation comports with recent studies identifying PPI or other gastric acid suppressant use as a significant risk factor for MDRGN carriage.Reference Huizinga, van den Bergh, van Rijen, Willemsen, van ‘t Veer and Kluytmans37, Reference D’Agata, Varu and Geffert38 Using these criteria for targeted surveillance would capture 21 of 217 colonized patients while producing only 3 false-positive screening referrals. Although recent CRO-positive cultures combined with either PPI usage or prior hospitalization were highly predictive, however, these criteria would still have missed 196 CRO-colonized patients (90%) who did not have these characteristics.
Interestingly, only 1 CPO-colonized patient had documented recent international hospitalization, the current CDC-recommended exposure for targeted CRE screening.15 Moreover, although CPO-colonized patients were significantly more likely than non-colonized patients to report foreign travel of themselves or a partner within the 21 preceding days, this variable did not emerge as a strong predictor in decision tree models (likely due to the few patients, only 0.6%, with this exposure).
This study highlights key challenges that may make predicting patients’ CRO/CPO colonization status, and in turn implementing successful targeted screening algorithms, difficult. First, although risk factors are important explanatory variables from an etiologic perspective and can identify where we may intervene to prevent an outcome, these variables reflect relative risk, not absolute risk. Risk factors are not necessarily good at predicting (ie, distinguishing between) who does or does not have an outcome, particularly when the number of affected patients is small. For example, although a recent CRO-positive culture was a strong risk factor (P < .001) for CRO colonization at admission, it only accounted for 34 of 217 cases. Eighty-four percent of CRO-colonized patients did not have a recent CRO-positive culture, and for the majority of our cohort, this variable would therefore not be helpful for predicting CRO status at admission. Second and similarly, high bacterial and genomic diversity among colonizing isolates may contribute to difficulty in predicting carriage by increasing outcome heterogeneity. In particular, CPOs reflected considerable organism and carbapenemase diversity, with one-third of CP-CREs encoding carbapenemases other than KPCs, including 2 genes (bla NDM and bla OXA-48-like) in a single organism. Third, although we collected extensive EMR data on healthcare-associated exposures, poor model sensitivity may reflect limitations of EMR data and not the absence of true predictive characteristics, particularly if colonization acquisition predated available retrospective periods. Finally, although risk factors for CRE and other CROs in US patients have traditionally focused on healthcare settings, increasing reports describe community reservoirs of carbapenem resistance (eg, porcine farms, retail seafood).Reference Mollenkopf, Stull and Mathys39, Reference Janecko, Martz and Avery40 These non-traditional exposures are unlikely to be systematically captured in the EMR.
Notwithstanding these challenges, our results offer actionable information. Recent CRO- or CRE-positive culture was consistently the strongest predictor of admission colonization, and many infection control programs already capture and flag these cultures. Moreover, 24% and 33% of CRO- and CPO-colonized patients, respectively, were co-colonized with VRE detected during routine admission screening. These patients would be placed on contact isolation precautions even without dedicated CRO surveillance. These findings suggest that existing screening policies may have unrecognized benefits and may justify continued surveillance and/or contact precautions for endemic VRE colonization.Reference Martin, Bryant and Grogan41, Reference Morgan, Wenzel and Bearman42
Our study has several limitations. First, this was a single-center study, and although we internally validated our models, our results should be validated in other cohorts. Our results may not be generalizable to other, lower-risk hospitalized populations or higher-endemicity areas (eg, New York City). Second, concordance between phenotypic and molecular carbapenemase assays was lower than expected,Reference Pierce, Simner and Lonsway20 particularly for E. cloacae, and further whole genome sequencing is planned to clarify this discrepancy. Nevertheless, sensitivity analyses restricting the CPO outcome to molecularly confirmed isolates yielded similar findings. Third, despite gathering extensive demographic and clinical information, there was likely missing exposure data (eg, outpatient antibiotic use, data that does not interface across hospitals). Many exposures, however, were strongly associated with study outcomes, consistent with other published literature. More importantly, because the prediction models were designed to inform real-world screening decisions, their performance under the practical constraints of potentially incomplete EMR data is arguably relevant.
Overall, in this high-risk inpatient population, CRO and CPO carriage was infrequent but higher than previously published estimates, including from other US ICU populations. There was significant organism and resistance mechanism diversity. We molecularly identified carbapenemases in 7 different bacterial species, providing an important reminder that many GI-colonizing organisms can serve as carbapenemase gene reservoirs. Despite including many patient characteristics associated with colonization or infection in the literature, overall, neither our machine learning–derived models nor current CDC targeted screening criteria (ie, recent foreign hospitalization) were highly accurate in predicting whether patients were colonized at admission. An important goal of artificial intelligence and other machine learning applications in health care is to capitalize on ‘Big data,’ despite its imperfections, to improve patient outcomes. Our study has demonstrated that currently available EMR data did not meet these targets. We believe that this was attributable, in part, to high exposure and microbiological heterogeneity, raising questions about how useful targeted screening strategies will be to identify CRO-colonized patients. Our models did successfully identify certain patient subgroups with high probabilities of colonization, however, including those with a recent history of CRO-positive culture(s) who use PPIs. Expanding upon existing CDC criteria to include other high-risk sub-groups may be useful as efforts continue to optimize CRE and CRO screening policies.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2019.42.
Author ORCIDs
Katherine E Goodman, 0000-0003-2851-775X
Acknowledgments
We would like to thank Verna Scheeler, Michael Anderson, Dina Khamash, and Sean Thompson for their assistance with study coordination, and data collection and validation, as well as Belita Opene, Shawna Lewis, Yehudit Bergman, and Krizia Chambers for their work processing surveillance cultures. We would also like to thank members of the JHU Clinical Microbiology Laboratory staff for helping with collection of surveillance swabs for the study.
Financial support
This work was supported by the Centers for Disease Control and Prevention Epicenters Program (grant no. 1U54CK000447), the CDC MIND-Healthcare Program (grant no. 1U01CK000536), the Agency for Healthcare Research and Quality (AHRQ grant no. R36HS025089), and The Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases.
Conflicts of interest
Dr Milstone reports personal fees from Becton Dickinson Diagnostics. Dr Rock and Dr Maragakis report grant support from the Clorox Company. Dr Cosgrove reports personal fees from Novartis and Theravance. Dr Tamma reports grants from Merck. All of these occurred outside the scope of the submitted work. Dr Simner reports grants and personal fees from Accelerate Diagnostics, grants and personal fees from Opgen, grants from BD Diagnostics, grants from bioMerieux, grants from Check-Points Diagnostics, grants from Hardy Diagnostics, personal fees from Roche Diagnostics, and personal fees from Oxford Nanopore, all outside the scope of the submitted work. All other authors report no potential conflicts of interest.