Introduction
The genus Lecithostaphylus currently comprises 11 species of trematodes parasitizing the intestines of marine fish from several orders. The genus is cosmopolitan, and the range of its definitive hosts is fairly broad (Zhang et al., Reference Zhang, Qiu and Li1986; Cribb et al., Reference Cribb, Bray and Barker1992; Toman, Reference Toman1992; Ramadan et al., Reference Ramadan, Morsy and Lashein2003; Châari et al., Reference Châari, Derbel and Neifar2013; Cabañas-Granillo et al., Reference Cabañas-Granillo, Solórzano-García, Mendoza-Garfias and Pérez-Ponce de León2020). The East Asian territory is the type region for a single species of this genus, Lecithostaphylus fugus Zhang, Qiu & Li, 1986, detected from tetraodontids in China (Zhang et al., Reference Zhang, Qiu and Li1986). The nearest localities from which Lecithostaphylus spp. have been reported are the Indian Ocean for Lecithostaphylus pomacentri Toman, 1992, Pacific waters near Australia for Lecithostaphylus gibsoni Cribb, Bray, & Barker, 1992 and the Hawaiian Islands for Lecithostaphylus depauperati Yamaguti, 1970.
Bray (Reference Bray, Bray, Gibson and Jones2008) considered Lecithostaphylus as a valid genus of the subfamily Lepidophyllinae Stossich, 1904. Later, several complex studies indicated that Lecithostaphylus was phylogenetically distant from the genus Lepidophyllum Odhner, 1902 and could be separated into a distinct subfamily Lecithostaphylinae Odhner, 1911 (Cabañas-Granillo et al., Reference Cabañas-Granillo, Solórzano-García, Mendoza-Garfias and Pérez-Ponce de León2020; Sokolov et al., Reference Sokolov, Shchenkov and Gordeev2021a; Reference Sokolov, Shchenkov, Gordeev and Ryazanova2021b). On the basis of molecular-based species clustering, Sokolov et al. (Reference Sokolov, Shchenkov, Gordeev and Ryazanova2021b) resurrected the subfamily Lecithostaphylinae and provided a diagnosis of Lecithostaphylinae sensu lato, including the genera Lecithostaphylus (as the type genus), Deretrema Linton, 1910, Proctophantases Odhner, 1911 and Steganoderma Stafford, 1904.
The genus Gymnotergestia Nahhas & Cable, 1964 (Fellodistomidae Nicoll, 1909) contains a single species, Gymnotergestia chaetodipteri Nahhas & Cable, 1964, described from the perciform fish Chaetodipterus faber (Broussonet, 1782) from the Caribbean Sea (Nahhas & Cable, Reference Nahhas and Cable1964). This species was also detected from Serranus scriba (Linnaeus, 1758) in the Mediterranean Sea off Libya (Al-Bassel, Reference Al-Bassel1999). There is a report of Gymnotergestia sp. in beloniform fish Arrhamphus sclerolepis Günther, 1866 from Australian coastal waters, and a drawing of this trematode is provided in the Keys to the Trematoda (Bray, Reference Bray, Bray, Gibson and Jones2002, p. 291). However, morphological description and metric data for these worms are absent.
At present, the structure of the families Zoogonidae and Fellodistomatidae and the species differentiation within Lecithostaphylus and Gymnotergestia are mainly based on morphological data. Though there are several molecular studies of trematodes from each of these two families (Bray et al., Reference Bray, Littlewood, Herniou, Williams and Henderson1999; Hall et al., Reference Hall, Cribb and Baker1999; Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003; Cribb et al., Reference Cribb, Miller, Bray and Cutmore2014, Reference Cribb, Bray, Hall and Cutmore2015; Cutmore et al., Reference Cutmore, Miller, Bray and Cribb2014; Antar & Gargouri, Reference Antar and Gargouri2015; Sokolov et al., Reference Sokolov, Gordeev and Lebedeva2016; Wee et al., Reference Wee, Cribb, Bray and Cutmore2017; Cutmore et al., Reference Cutmore, Bray and Cribb2018; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Anglade and Randhawa2018; Cabañas-Granillo et al., Reference Cabañas-Granillo, Solórzano-García, Mendoza-Garfias and Pérez-Ponce de León2020; Krupenko et al., Reference Krupenko, Uryadova, Gonchar, Kremnev and Krapivin2020; Sokolov et al., Reference Sokolov, Shchenkov and Gordeev2021a, Reference Sokolov, Shchenkov, Gordeev and Ryazanovab), most of them do not provide morphological validation of the species under study. In many cases, this means that no sound taxonomical conclusions can be made.
During our survey of the trematode fauna of fish in the coastal waters of northern Vietnam, we found mature trematode specimens morphologically similar to Lecithostaphylus Odhner, 1911 (Zoogonidae Odhner, 1902: Lepidophyllinae Stossich, 1903) and Gymnotergestia Nahhas & Cable, 1964 (Fellodistomidae Nicoll, 1909: Tergestiinae Skrjabin & Koval, 1957) in the intestines of Hemiramphus Cuvier, 1816 and Strongylura van Hasselt, 1824, respectively. In this study, we describe two new trematode species based on the results of morphological and molecular examination of these worms.
Materials and methods
Collection of trematodes
Specimens of trematodes from the families Zoogonidae and Fellodistomidae were sampled from four individuals of Hemiramphus far (Forsskål, 1775), from one out of two individuals of Hemiramphus marginatus (Forsskål, 1775), and from ten out of 18 individuals of Strongylura strongylura (van Hasselt, 1823) in the coastal waters of Cat Ba Island, northern Vietnam (20°84′N, 106°59′E). The worms were briefly rinsed in distilled water and killed in hot distilled water. Several specimens were preserved in 70% ethanol for preparation of slides, while others were fixed in 96% ethanol for DNA extraction. Whole-mounts for the descriptions of adult worms were made by staining them with alum carmine, dehydrating them in a graded ethanol series, clearing in xylene and mounting in Canada balsam under a coverslip on a slide. All measurements are given in micrometres.
DNA extraction, amplification and sequencing
Adult specimens of Lecithostaphylus halongi n. sp. (n = 2) and Gymnotergestia strongyluri n. sp. (n = 2) were used for molecular analysis (table 1). Total DNA was extracted from the worms fixed in 96% ethanol using a ‘hot shot’ technique (Truett, Reference Truett and Kieleczawa2006).
Table 1. List of taxa incorporated in the molecular analysis of the superfamily Microphalloidea with the number of 28S rDNA sequences given in parentheses.

28S ribosomal DNA (rDNA) was amplified using 28S-A forward primer (5′-GCACCCGCTGAAYTTAAG-3′) (Matejusova & Cunningham, Reference Matejusova and Cunningham2004) and 1500R (5′-GCTATCCTGAGGGAAACTTCG-3′) (Tkach et al., Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003). Initial polymerase chain reaction (PCR) reaction was performed in a total volume of 25 μl containing 0.25 mm of each primer pair, 25 ng of total DNA in water and 12.5 μl of Promega GoTaq Green Master mix (Madison, Wisconsin, USA). Amplification of a 1200-bp fragment of 28S ribosomal RNA (rRNA) gene was performed in a GeneAmp 9700 (Applied Biosystems, Waltham, Massachusetts, USA), with a 5-min denaturation at 96°C, 35 cycles of 1 min at 96°C, 20 s at 55°C and 2 min 30 s at 72°C, and a 10-min extension at 72°C. Negative and positive controls were made with the use of both primers.
PCR products were directly sequenced using an ABI Big Dye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, Massachusetts, USA), as recommended by the manufacturer, with the internal sequencing primers described by Tkach et al. (Reference Tkach, Littlewood, Olson, Kinsella and Swiderski2003) for 28S rDNA. PCR products were analysed using an ABI 3500 genetic analyser at the Federal Scientific Center of the East Asia Terrestrial Biodiversity FEB RAS. The sequences were submitted to the National Center for Biotechnology Information (NCBI) database with accession number: (OK636406 - OK636409).
Alignment and phylogenetic analysis
rDNA sequences were assembled with SeqScape v.2.6 software (provided by Applied Biosystems, Waltham, Massachusetts, USA). Alignments and estimation of the number of variable sites and sequence differences were performed using MEGA 7.1 (Kumar et al., Reference Kumar, Stecher and Tamura2016).
Phylogenetic analysis of nucleotide sequences was undertaken using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. Prior to the analysis, the nucleotide substitution model was estimated using Akaike's information criterion for ML (Akaike, Reference Akaike1974) and Bayesian information criterion (BIC) for BI (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001) using jModeltest v.3.07 software (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). The model TVM + I + G (Posada, Reference Posada2003) was estimated as best fitting the 28S rDNA sequence data of Zoogonidae for both ML and BI analyses. The models GTR + I+G and TPM3uf + G (Posada, Reference Posada2003) were estimated as optimal for 28S rDNA of the Fellodistomidae dataset for ML and BI algorithms, respectively. Phylogenetic trees were reconstructed with PhyML 3.1 (Guindon & Gascuel, Reference Guindon and Gascuel2003) and MrBayes v.3.1.2 software (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001). A Bayesian algorithm was performed using the Markov Chain Monte Carlo option with ngen = 10,000,000, nruns = 2, nchains = 4, temp = 0.5 and samplefreq = 100. Burn-in values for ‘sump’ and ‘sumt’ options made up 25% of the number of generations (ngen). Phylogenetic relationship significance was estimated using posterior probabilities for both ML and BI analyses (Huelsenbeck et al., Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001).
Phylogenetic relationships were inferred from our data and the nucleotide sequences of 28S rDNA from other trematode specimens from the superfamilies Microphalloidea and Gymnophalloidea obtained from the NCBI GenBank database (tables 1 and 2).
Table 2. List of taxa incorporated in the molecular analysis of the family Fellodistomidae with the number of 28S rDNA sequences given in parentheses.

Results
Lecithostaphylus halongi n. sp.
Taxonomic summary
-
Type host. Hemiramphus far (Forsskål, 1775).
-
Other host. Hemiramphus marginatus (Forsskål, 1775).
-
Site. Intestine.
-
Intensity of infection. 1–5 specimens.
-
Type locality. Coastal water of Cat Ba Island, Tonkin Bay, northern Vietnam (20°84′N, 106°59′E).
-
Type deposition. Holotype no. 194–Tr, paratypes nos. 195–198–Tr.
-
Materials deposited. Materials are deposited in the parasitological collection of the Zoological Museum (deposited 20 November 2020, Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far East Branch of the Russian Academy of Sciences, Vladivostok, Russia; e-mail: petrova@biosoil.ru).
-
Etymology. Species name refers to Halong Bay, Vietnam, where the fish infected with this parasite were caught.
Description
Adult. Based on five specimens (table 3 and fig. 1a). Body broadly fusiform. Tegumental spines lacking. Oral sucker globular, subterminal. Ventral sucker round, larger than oral sucker, pre-equatorial. Prepharynx short. Pharynx round. Oesophagus extremely short, bifurcates immediately anteriorly to ventral sucker. Caeca extend to posterior third of hindbody. Testes round to oval, symmetric, separated by uterus, located just anteriorly of border between anterior and posterior part of hindbody. Cirrus sac elongate, curved in posterior part, dorsally to ventral sucker, dextral to mid-body line, extending from posterior end of ventral sucker to genital pore. Genital pore sinistral, lateral at level of posterior half of pharynx. Seminal vesicle elongate, occupies posterior third of cirrus sac. Pars prostatica tubular, surrounded by few prostatic cells. Ejaculatory duct long. Ovary submedian, transversally oval, partially overlapping ventral sucker. Seminal receptacle median, in space between ovary and testes. Vitellarium in two clusters of 9–10 round follicles, extending from level of posterior border of ventral sucker or from level of ovary to middle or posterior third of testes. Uterus mainly in post-testicular region. Metraterm long, extending from mid-level of ventral sucker to genital pore. Eggs numerous, operculate. Excretory vesicle not observed, excretory pore terminal.
Table 3. Measurements (μm) of adult worms Zoogonidae (Microphalloidea) and Fellodistomidae (Gymnophalloidea).

Bw, Body width; Bl, Body length; Fo, Forebody.
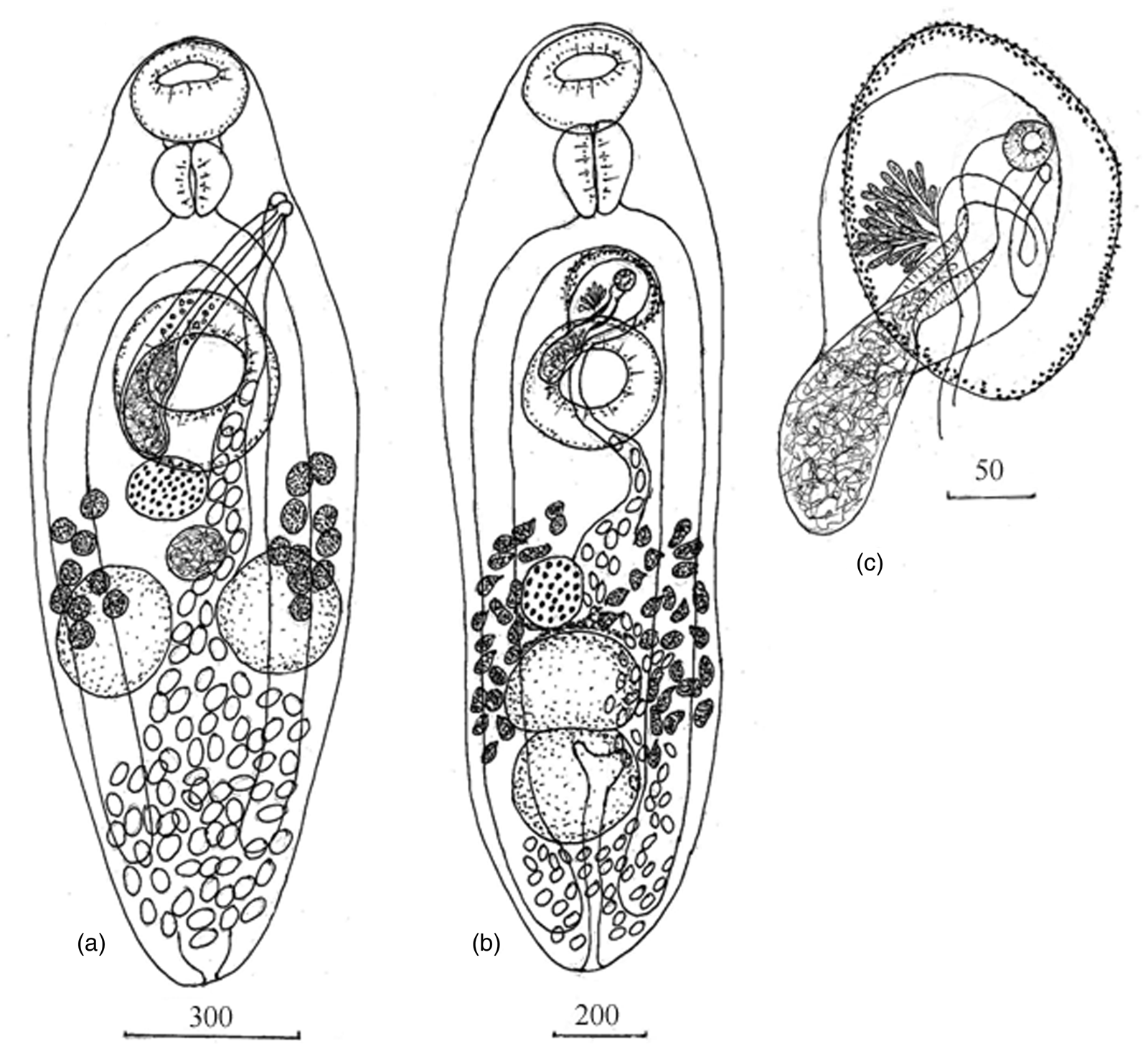
Fig. 1. Adult worms: (a) Lecithostaphylus halongi n. sp., ventral view; (b, c) Gymnotergestia strongyluri n. sp., ventral view. Measurements are given in μm.
Molecular data
Partial sequences of 28S rRNA gene 1164 bp in length of two specimens of L. halongi n. sp. (OK636406-OK636407) were aligned with all available ribosomal large subunit sequences of the Zoogonidae and the Faustulidae and trimmed to the most optimal alignment length (1105 bp) for the available dataset; missing data were considered for the new species when building the alignment. The two 28S rDNA sequences of L. halongi n. sp. obtained in our study were identical.
Gymnotergestia strongyluri n. sp.
Taxonomic summary
-
Type host. Strongylura strongylura (van Hasselt, 1823).
-
Site. Intestine.
-
Intensity of infection. 1–26 specimens.
-
Type locality. Coastal water of Cat Ba Island, Tonkin Bay, northern Vietnam (20°84′N, 106°59′E).
-
Type deposition. Holotype no. 199–Tr, paratypes nos. 200–203–Tr.
-
Materials deposited. Materials are deposited in the parasitological collection of the Zoological Museum (deposited 20 November 2018, Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far East Branch of the Russian Academy of Sciences, Vladivostok, Russia; e-mail: petrova@biosoil.ru).
-
Etymology. The species name refers to the definitive host, S. strongylura.
Description
Adult. Based on five specimens (table 3 and fig. 1b, c). Body elongate oval. Tegumental spines lacking. Oral sucker globular, subterminal. Ventral sucker round, larger than oral sucker, pre-equatorial. Prepharynx not observed. Pharynx large, elongate, conical, expanding anteriorly and tapering posteriorly. Oesophagus extremely short, bifurcates at midway between oral and ventral sucker. Caeca slightly not reaching posterior end of body. Testes round to oval, tandem, adjacent to each other, located in posterior third of body. Cirrus sac pyriform with expanding anterior part located immediately anteriorly to ventral sucker and elongate posterior part at level of anterior half of ventral sucker. Genital pore with muscular sphincter, sinistral, submedian between intestinal bifurcation and ventral sucker, opening into genital atrium. Genital atrium oval, surrounded by numerous small glandular cells. Seminal vesicle elongate, occupies elongate part of cirrus sac and partly penetrates expanding part of cirrus sac. Pars prostatica thick-walled, surrounded by numerous prostatic cells. Ejaculatory duct curved. Ovary round or oval, submedian, immediately before testes. Vitellarium in two lateral fields of irregular follicles. Vitelline fields extending from level of middle of distance between ovary and ventral sucker to anterior half of posterior testis. Uterus occupying much of hindbody and part of lateral region, sinistral to testes and ovary. Metraterm thin-walled, long, curved, extending from posterior end of ventral sucker to genital pore. Eggs numerous, operculate. Excretory vesicle Y-shaped, pore terminal.
Molecular data
Partial sequences of 28S rRNA gene 1270 bp in length of two specimens of G. strongyluri n. sp. (OK636408-OK636409) were aligned with all available ribosomal large subunit sequences of the Fellodistomidae, resulting in a 908 bp alignment dataset. The two 28S rDNA sequences of G. strongyluri n. sp. differed by four variable sites, which represents 0.46 ± 0.21% of divergence.
Remarks
The worms found in Hemiramphus spp. from Vietnam apparently belong to the genus Lecithostaphylus and are morphologically similar to L. gibsoni Cribb, Bray & Barker, 1992 ex Abudefduf whitleyi Allen & Robertson, 1974 (Pomacentridae) of Heron Island (Cribb et al., Reference Cribb, Bray and Barker1992) and to L. depauperati Yamaguti, 1970 ex Hemiramphus depauperatus Lay & Bennett, 1839 off Hawaii (Yamaguti, Reference Yamaguti1970). There are no marked differences between the two trematodes described in this study and the two known species just mentioned in the metric parameters of the body and the eggs and the size of most organs, except the length of the cirrus sac (table 3). The cirrus sac of trematodes from our study is longer than that of L. gibsoni and L. depauperati. The worms collected in Vietnam also differ from L. gibsoni and L. depauperati by organ topology. In trematodes ex Hemiramphus spp., vitelline fields extend from the level of the posterior border of the ventral sucker or ovary to the middle or posterior third of the testes, while in L. gibsoni, vitelline fields are predominantly located anteriorly to the testes at the level of the ventral sucker, and in L. depauperati, vitelline fields reach the anterior border of the ventral sucker. On the basis of these data, we recognize the worms ex Hemiramphus spp. from Vietnam as a new species and name it L. halongi n. sp.
To check the validity of the new species, ML and BI phylogenetic analyses based on 28S rDNA partial sequences were performed for the Zoogonidae. These two algorithms generated phylogenetic trees with an identical topology (fig. 2). We omitted the gymnophalloid clade of the Zoogonidae because its species always clustered with the Gymnophalloidea in previous molecular phylogenetic analyses (Sun et al., Reference Sun, Bray, Yong, Cutmore and Cribb2014; Cutmore et al., Reference Cutmore, Bray and Cribb2018; Diaz, Reference Diaz2018; Pérez-Ponce de León & Hernández-Mena, Reference Pérez-Ponce de León and Hernández-Mena2019; Sokolov et al., Reference Sokolov, Shchenkov and Gordeev2021a).

Fig. 2. Bayesian phylogenetic tree of Zoogonidae based on the analysis of partial 28S rRNA gene sequences; nodal numbers indicate posterior probabilities. Sequences from the present study are in bold. *Morphologically validated species. Scale bar shows the number of substitutions per site.
The results of the phylogenetic analysis indicate that L. halongi n. sp. is closely related to Lecithostaphylus brayi Cabañas-Granillo, Solórzano-García, Mendoza-Garfias & Pérez-Ponce de León, 2020 with a divergence value of 7.14 ± 0.91%, generating a monophyletic highly supported clade. Deretrema spp. and Proctophantastes spp. formed a distinct clade, a sister to Lecithostaphylus, with a high statistical support and a genetic divergence of 17.08 ± 0.98%. For comparison, the interspecific genetic divergence value for the monophyletic genus Lepidophyllum in our study was 14.76 ± 1.12%, which is similar to the divergence values between the different zoogonid genera on the phylogenetic tree. These results unambiguously indicate that the new species is a member of the genus Lecithostaphylus within the subfamily Lecithostaphylinae, which also includes Deretrema nahaense Yamaguti, 1942 and Proctophantastes gillissi (Overstreet & Pritchard, 1977) Bray & Gibson, 1986 from the present dataset.
We disagree with the opinion of Sokolov et al. (Reference Sokolov, Shchenkov and Gordeev2021a) about the existence of Lecithostaphylinae sensu lato, which includes the species mentioned above and Steganoderma eamiqtrema Blend & Racz, 2020. Steganoderma eamiqtrema has high p-distance values, 20.44 ± 1.21%–23.68 ± 1.23%, as compared with species of Lecithostaphylinae sensu stricto by 28S rDNA sequence data, and the phylogenetic relationships of this species are poorly supported in the study by Sokolov et al. (Reference Sokolov, Shchenkov, Gordeev and Ryazanova2021b). Moreover, in our study, S. eamiqtrema (Zoogonidae) was sister to [Lecithostapyhllinae + Zoogoninae + Faustulidae (Antorchis pomacanthi, Trigonocryptus conus and Bacciger lesteri)] clade on both ML and BI trees with a high support (figs 2 and 3). In our opinion, the molecular data indicate that S. eamiqtrema belongs to a distinct subfamily. Thus, we suggest that Lecithostaphylus should be removed from the Lepidophyllinae and that the subfamily Lecithostaphylinae Odhner, 1911 should be recognized, including at least three genera: Lecithostaphylus, Proctophantastes and Deretrema.

Fig. 3. (a) Phylogenetic tree of the superfamily Gymnophalloidea based on the analysis of partial 28S rRNA gene sequences; nodal numbers indicate posterior probabilities for ML/BI algorithms. Sequences from the present study are in bold. (b) Fragment of ML phylogenetic tree topology exhibited relationships of Gymnotergestia strongyluri n. sp. Sequences from the present study are in bold. Scale bar shows the number of substitutions per site.
The diagnostic characters of trematode specimens found in S. strongylura from Vietnam correspond to those of Gymnotergestia. This genus contains a single species, G. chaetodipteri Nahhas & Cable, 1964, ex C. faber from Jamaica (Nahhas & Cable, Reference Nahhas and Cable1964). The worms from our material are similar to G. chaetodipteri in having a conical pharynx, tandem testes, a pre-testicular ovary, vitelline follicles in lateral fields posterior to the ventral sucker and in not having external seminal vesicle and seminal receptacle. At the same time, the worms ex S. strongylura, unlike G. chaetodipteri, possess an elongate to oval body vs. elongate slender body, and testes adjacent to each other vs. testes separated by uterine coils. Besides, there are considerable differences in the pharynx length, egg size and the suckers ratios between these two species (table 3).
Taken together, these data justify the establishment of a new species, G. strongyluri n. sp., for the worms described in our study. Morphologically, they are most similar to the specimens of Gymnotergestia sp. ex A. sclerolepis from Australia in the shape of the cirrus sac and the relative arrangement of testes and ovary, on the basis of the figure provided in Keys to the Trematoda (Bray, Reference Bray, Bray, Gibson and Jones2002 p. 291). However, these worms differ from each other by the cirrus sac and vitellarium arrangement relative to other organs. Unfortunately, there are no published morphometric data for Australian worms, but judging from the figure in Bray (Reference Bray, Bray, Gibson and Jones2002), they may belong to a new species of Gymnotergestia.
It is also impossible to perform a meaningful comparative analysis of the worms from our material and those from the study of Al-Bassel (Reference Al-Bassel1999), who found G. chaetodipteri in S. scriba from the Mediterranean Sea. The illustration provided in Al-Bassel (Reference Al-Bassel1999) is poorly informative for taxonomical interpretations, and molecular data are needed for any conclusion.
Phylogenetic trees of the Fellodistomatidae based on 28S rDNA have identical topologies in ML and BI analyses, except relationships within the clade containing G. strongyluri n. sp. and the genus Tergestia Stossich, 1899 (fig. 3a, b). These relationships appear as a poorly supported polytomy of G. strongyluri n. sp. and Tergestia spp. (except Tergestia henryi Wee, Cutmore, Yong & Cribb, 2017) on the Bayesian tree (fig. 3a) and as a poorly supported dichotomy on the ML tree (fig. 3b), indicating paraphyly of Tergestia. Genetic divergence by 28S rDNA sequence data of G. strongyluri n. sp. and Tergestia spp. ranged from 4.61 ± 0.68% to 7.48 ± 0.81%, which was comparable with that for interspecific divergence values within Tergestia (1.85 ± 0.44 to 9.09 ± 0.94%) and for two other fellodistomid genera: Steringophorus Odhner, 1905 (1.61 ± 0.39 to 7.59 ± 0.86%) and Proctoeces Odhner, 1911 (4.26 ± 0.71 to 8.9 ± 0.98%). For the latter genus, a minimal value of 4.26 ± 0.71% was observed for different specimens of Proctoeces maculatus (Looss, 1901) Odhner, 1911 ex Archosargus probatocephalus (Walbaum, 1792), Gulf of Mexico, Mississippi, USA (Olson et al., Reference Olson, Cribb, Tkach, Bray and Littlewood2003) and ex Sabella pavonina Savigny, 1822, Bizerte Lagoon, Tunisia (Antar & Gargouri, Reference Antar and Gargouri2015). Antar & Gargouri (Reference Antar and Gargouri2015) provided conclusive morphological and molecular evidence that the specimen of P. maculatus ex A. probatocephalus belongs to a different species of Proctoeces. Thus, we can use this value as reliable criterion for the minimal broad interspecific genetic divergence between Proctoeces species.
In our study, G. strongyluri n. sp. and Tergestia spp. clustered together on the phylogenetic tree, differing from each other by the p-distance values at the interspecific level. This indicates that these trematodes may belong to the same genus. However, additional molecular data for most of the species of these two genera, especially the type species, are needed for a final taxonomical conclusion.
In our analysis, we did not consider the genetic divergence between Olssonium turneri Bray & Gibson, 1980 and Fellodistomum spp., which ranged from 2.41 ± 0.5% to 2.52 ± 0.52% by 28S rDNA sequences. These values correspond to the interspecific divergence level. However, this fact was ignored by all the authors of the phylogenetic studies of the Fellodistomidae employing a molecular approach, even though relatively representative data were available (Bray et al., Reference Bray, Littlewood, Herniou, Williams and Henderson1999; Cribb et al., Reference Cribb, Miller, Bray and Cutmore2014; Antar & Gargouri, Reference Antar and Gargouri2015; Wee et al., Reference Wee, Cribb, Bray and Cutmore2017; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Anglade and Randhawa2018; Krupenko et al., Reference Krupenko, Uryadova, Gonchar, Kremnev and Krapivin2020). In our opinion, it should be checked in further complex studies whether O. turneri in fact belongs to the genus Fellodistomum.
Financial support
This study was funded by the federal budget of the Russian Academy of Sciences (project no. 0228-2019-0002).
Conflicts of interests
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional guides on the care and use of laboratory animals.








