Introduction
Siphonaptera is relatively a small order of secondarily wingless holometabolous insects. According to Beaucournu & Gomez-Lopez (Reference Beaucournu and Gomez-Lopez2015) the order includes 2500 species ‘of fleas’. In addition, 409 specific, 147 subspecific, 65 generic, and 7 subgenera names are considered to be synonymous (Krasnov, Reference Krasnov2008). The Siphonaptera fauna of the Palearctic region is the richest, including 96 genera and 892 species constituting a 38% of the total number of species known, and 38% of the known genera (Krasnov, Reference Krasnov2008).
Within this order, the Pulicidae is the most studied family since most fleas of medical or veterinary importance (Ctenocephalides felis, Ctenocephalides canis, Pulex irritans or Xenopsylla cheopis) are members of this family. Pulicidae consists of 4 tribes, 21 genera, and 167 species. Some workers have treated Pulicidae as including Tungidae (Lewis, Reference Lewis1998); however, Whiting et al. (Reference Whiting, Whiting, Hastriter and Dittmar2008) placed this family as a monophyletic group and phylogenetically distant from Tungidae. Pulicidae exhibit an interesting diversity of host specificity patterns and ecological habits (Whiting et al., Reference Whiting, Whiting, Hastriter and Dittmar2008). Certain species such as Archaeopsylla erinacei and Spilopsyllus cuniculi are monoxenous on hedgehog and rabbits respectively, while other Pulicidae species such as C. felis or P. irritans, are highly promiscuous, and occurs on a wide variety of Carnivora (Whiting et al., Reference Whiting, Whiting, Hastriter and Dittmar2008).
Although during the last 15 years molecular data has made a significant contribution (Dittmar & Whiting, Reference Dittmar and Whiting2003; Vobis et al., Reference Vobis, D'Haese, Mehlhorn, Mencke, Blagburn, Bond, Denholm, Dryden, Payne, Rust, Schroeder, Vaughn and Bledsoe2004; Gamerschlag et al., Reference Gamerschlag, Mehlhorn, Heukelbach, Feldmeier and D'Haese2008; Whiting et al., Reference Whiting, Whiting, Hastriter and Dittmar2008; Marrugal et al., Reference Marrugal, Callejón, de Rojas, Halajian and Cutillas2013; Zurita et al., Reference Zurita, Callejón, De Rojas, Halajian and Cutillas2016), for decades, the genus and species differentiation of fleas has been based on morphological criteria (the shape and structure of their complex genitalia, distribution of setae, spines, and ctenidia, etc) (Lane & Crosskey, Reference Lane and Crosskey1993; Kramer & Mencke, Reference Kramer and Mencke2001; Mehlhorn, Reference Mehlhorn2001; Linardi & Santos, Reference Linardi and Santos2012). However, a few studies have been carried out on the molecular differentiation of fleas (Lawrence et al., Reference Lawrence, Brown, Peters, Spielman, Morin-Adeline and Slapeta2014; Zurita et al., Reference Zurita, Callejón, De Rojas, Gómez-López and Cutillas2015). Thus, the scientific community has a great knowledge of flea taxonomy at the species and subspecies level, and enough information to assess their biology and role in disease transmission in recent years (Kaewmongkol et al., Reference Kaewmongkol, Kaewmongkol, Mclnnes, Burmej, Bennet, Adams, Ryan, Irwin and Fenwick2011; Lawrence et al., Reference Lawrence, Hii, Jirsová, Panáková, Ionica, Gilchrist, Modrý, Mihalca, Webb, Traub and Slapeta2015). In contrast, a rigorous exploration of the phylogenetic relationships among fleas is needed in order to clarify their complex systematics (Whiting et al., Reference Whiting, Whiting, Hastriter and Dittmar2008). In this way, the few taxonomic and phylogenetic studies of fleas based on molecular data carried out in the last years have revealed that not all flea species previously described only by morphological methods have always remained as defined species. Recently, Zurita et al. (Reference Zurita, Callejón, De Rojas and Cutillas2018) based on a comparative morphological, phylogenetic and molecular study of Nosopsyllus fasciatus and Nosopsyllus barbarus, concluded that there were no solid arguments to consider these two ‘morphospecies’ as two different species and proposed N. barbarus as a junior synonym of N. fasciatus. These authors used two nuclear markers: Internal Transcribed Spacers 1 and 2 (ITS1 and ITS2) and two mitochondrial markers: cytochrome c-oxidase subunit 1 (cox1) and cytochrome b (cytb), in order to determine the taxonomic status of both species.
Previous studies showed that fleas have a high level of genetic intraspecific variation (Dittmar & Whiting, Reference Dittmar and Whiting2003; Brinkerhoff et al., Reference Brinkerhoff, Martin, Jones and Collinge2011). Thus, several authors in the last 10 years (Kaewmongkol et al., Reference Kaewmongkol, Kaewmongkol, Mclnnes, Burmej, Bennet, Adams, Ryan, Irwin and Fenwick2011; Lawrence et al., Reference Lawrence, Brown, Peters, Spielman, Morin-Adeline and Slapeta2014; Zhu et al., Reference Zhu, Hastriter, Whiting and Dittmar2015; Zurita et al., Reference Zurita, Callejón, De Rojas, Gómez-López and Cutillas2015, Reference Zurita, Callejón, De Rojas, Halajian and Cutillas2016) have used mitochondrial DNA markers such as cox1, coxII or cytb as reference molecular markers in order to investigate the phylogenetic and taxonomic relationships in fleas at family, genus and species level.
Genus Archaeopsylla Dampf, 1908 (Pulicidae) is a great example of the shortage of molecular and phylogenetic data in fleas’ taxonomy. Based on morphological criteria, two species have been described within the Archaeopsylla genus (Pulicidae): Archaeopsylla sinensis, and A. erinacei with two subspecies: Archaeopsylla erinacei erinacei (Bouché, Reference Bouché1835), and Archaeopsylla erinacei maura (Jordan & Rothschild, 1911). Both species have a Palearctic distribution; however, A. sinensis occurs at East-Asian subregion, Siberian province; China, Russia (Medvedev et al., Reference Medvedev, Lobanov and Lyangouzov2005) whereas A. erinacei is distributed from European region to Mediterranean and North Africa area (Hopkins & Rothschild, Reference Hopkins and Rothschild1953). Furthermore, A. e. erinacei is distributed from European and Mediterranean subregions, while the distribution of A. e. maura is possibly partly accounted by the artificial introduction of its host (the North African hedgehog, Atelerix algirus), primarily a North African form, which is stated to have, probably, been introduced into southern Spain (Domínguez, Reference Domínguez2004), the Balearic Islands and south-eastern France within historic times (Hopkins & Rothschild, Reference Hopkins and Rothschild1953). Thus, we can say that both subspecies are sympatric along certain geographical areas where they coexist and particularly also in the Iberian Peninsula and south-eastern France. Furthermore, Hopkins & Rothschild (Reference Hopkins and Rothschild1953) and Beaucournu & Launay (Reference Beaucournu and Launay1990) noticed that these two subspecies cohabit the same host (Erinaceus europaeus). These authors provided taxonomic keys based on morphological criteria in order to discriminate between the two subspecies; however, the close likeness of female specimens of A. e. erinacei and A. e. maura makes the differential diagnosis very difficult, especially when there are few males (easily differentiated), and when the specimens come from areas where the two subspecies coexist (Beaucournu & Launay, Reference Beaucournu and Launay1990).
The aim of this study was to carry out a comparative morphological, biometrical and molecular study of A. erinacei and their subspecies: A. e. erinacei and A. e. maura isolated from Erinaceus europaeus from Seville (southwestern of Spain) and Corse Island (France). Thus, the partial 18S rRNA gene, ITS1, ITS2 of the rDNA and partial cox1 and cytb mtDNA gene of these taxa were sequenced in order to clarify their taxonomic status and to assess intra-specific and intra-population similarity. Furthermore, based on the sequences obtained and those of additional flea species retrieved from public databases, we also carried out a comparative phylogenetic analysis.
Materials and methods
Collection of samples
Hedgehog (Erinaceus europaeus) trapping was conducted in Dos Hermanas (37°17′01″N–5°55′20″W) and Aznalcázar (37°18′14″N–6°15′03″W), Seville (Spain). Early morning hedgehogs killed on roads at night were located and collected by hand. Collected hedgehogs were taken to the laboratory and then placed on a white sheet of paper in order to be visually examined for ectoparasites. Fleas were collected by adding 70% ethanol and then were removed from the hedgehogs by gently shaking the animal over the white sheet of paper. Fleas from hedgehog from Corse were obtained through the assistance of colleagues (see Acknowledgements). Fleas obtained were kept in Eppendorf tubes with 70% ethanol until required for subsequent identification and sequencing; for details on locality, host, flea species and gender, see table 1.
Table 1. GenBank accession numbers of ITS1, ITS2 and cytb, cox1 and partial 18S gene sequences of individuals of A. erinacei and X. cheopis obtained in this study.

Morphological identification and biometrical study
Flea specimens collected from Spain were classified by us whereas those fleas from Corse provided by our colleagues were classified firstly by them (see Acknowledgements) and then compared morphologically with our specimens in our laboratory. For morphological analysis, all specimens were examined and photographed under an optical microscope. Posteriorly, flea legs were cut off in order to carry out the DNA extraction, while the rest of the flea was used to confirm A. erinacei species/subspecies morphological identity. Thus, they were cleared with 10% KOH, prepared and mounted on glass slides using conventional procedures (Lewis, Reference Lewis1993). Once mounted, they were examined and photographed again for a deeper morphological analysis using a Nikon microscope equipped with a camera lucid system and a photomicroscope. Generic, specific and subspecific identification was carried out according to Jordan & Rothschild (Reference Jordan and Rothschild1912), Hopkings & Rothschild (Reference Hopkins and Rothschild1953) and Beaucournu & Launay (Reference Beaucournu and Launay1990). Thus, the morphological characteristics considered for the specific determination include:
• Presence of a well noticed sclerotized falx of head.
• Asymmetrical antenna with partially welded basal segments.
• Presence of a pleural rod of mesothorax.
• Vestigial genal and pronotal comb. Genal comb composed of one to three spines, these being the small posterior ones. Pronotal comb composed of at most six spines on the two sides together, and sometimes only one each side. Very rarely some of these combs are entirely absent, but it can occur.
• Hind tibia with six seta-bearing notches along dorsal margin with a row of six to eleven little setae near to dorsal margin.
For the subspecific differentiation, we considered morphological characteristics reported by Jordan & Rothschild (Reference Jordan and Rothschild1912), Hopkings & Rothschild (Reference Hopkins and Rothschild1953)) and Beaucournu & Launay (Reference Beaucournu and Launay1990):
• Male specimens of A. e. erinacei showed the greatest length of basimere same as distance from base of spine on genal process to anterior edge of eye while, male individuals of A. e. maura showed the greatest length of basimere same as distance from base of spine at tip of genal process to front margin on head.
• Females of A. e. erinacei showed eighth abdominal tergum bearing two lateral bristles towards the base and seventh sternum usually with five lateral bristles on the two sides together, whereas A. e. maura females presented only one bristle in eighth abdominal tergum and seventh sternum usually bore four lateral bristles on the two sides together.
Furthermore, 20 different parameters were measured of 48 (23 females and 25 males) A. erinacei specimens (table 2). Descriptive univariate statistics (arithmetic means, standard deviations, and variation coefficients) for all parameters were determined for two populations (A. erinacei from Seville and A. erinacei from Corse) using IBM® SPSS® Statistics program version 24.0.0.0 (Pardo & Ruiz, Reference Pardo and Ruiz2002). In addition, a two-way Analysis of Variance (ANOVA), with a factorial design, was used in order to test the significance of the differences between geographic origin (GO) and sex. Different individuals of each population corresponding with the number of replications were considered for each combination of sex and GO. Means were compared using the Fisher's Least Significant Difference (LSD). The effect of GO, sex (S) and the interaction (GO × S) were calculated as the fraction of the total variability explained. All data analysis was performed with the software ‘Statistix 9.0’. Statistically significant differences were assumed for P < 0.05 (*).
Table 2. Biometrical data of A. erinacei specimens collected from hedgehogs from Seville and Corse.

TLF, total female length; TLM, total male length; TWF, total female width; TWM, total male width; HLF, total length of the female head; HLM, total length of the male head; HWF, total width of the female head; HWM, total width of the male head; BL, total length of the basimere; BW, total width of the basimere; GHL, Distance from base of spine at tip of genal process to front margin on head; GEL, distance from base of spine on genal process to anterior edge of eye; EL, total length of the spermatheca; EW, total width of the spermathecal; PL, total length of falx; DS7, distance among setae of seventh sternum; DSS, distance between more posterior setae of eighth abdominal tergum and posterior margin of the abdomen; PROLM, total male length of the prothorax; PROLF, total female length of the prothorax; MESLM, total male length of the mesothorax; MESLF, total female length of the mesothorax; METLM, total male length of the metathorax; METLF, total female length of the metathorax; MAX, maximum; MIN, minimum; б, standard deviation; X, arithmetic mean; VC, variation coefficient (%).
ANOVA TEST: Fisher's Least Significant Difference (LSD) and coefficient of variation (CV (%)) for each parameter.
*Mean significant differences (P < 0.05) according to LSD test.
Molecular study
The DNeasy Blood and Tissue Kit (Qiagen) extracted total DNA from flea legs according to the manufacturer's protocol. Then, genomic DNA was checked using an electrophoresis in 0.8% agarose gel electrophoresis infused with ethidium bromide.
All molecular markers sequenced in this study were amplified by polymerase chain reaction (PCR) using a thermal cycler (Eppendorf AG). PCR mix, PCR conditions, and PCR primers are summarized in table S1. The 18S, ITS1, ITS2, partial cox1, and cytb gene sequences obtained from A. erinacei from the two geographical areas were deposited in GenBank database (table 1). Furthermore, we sequenced and provided ITS2 and cytb sequences of Xenopsylla cheopis isolated from Rattus sp. from El Hierro Island (Spain) (see table 1).
The PCR products were checked on ethidium bromide stained 2% Tris–Borate–EDTA (TBE) agarose gels. Bands were eluted and purified from the agarose gel by using the QWizard SV Gel and PCR Clean-Up System Kit (Promega). Once purified, the products were sequenced by Stab Vida (Portugal). To obtain a nucleotide sequence alignment, we used MUSCLE alignment method (Edgar, Reference Edgar2004) by the MEGA program version 5.2 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011). The rDNA intra-individual variation was determined by sequencing 7–8 clones of one individual. The PCR products were eluted from the agarose gel using the WIZARD® SV Gel and PCR Clean-Up System (Promega) and the transformation was carried out as cited by Cutillas et al. (Reference Cutillas, Callejón, de Rojas, Tewes, Ubeda, Ariza and Guevara2009). Plasmids were purified using a Wizard Plus SV (Promega) and sequenced by Stab Vida (Portugal) with a universal primer (M13).
A restriction map of the ITS1 and ITS2 sequences of A. erinacei from Seville and Corse was constructed using The Sequence Manipulation Suite (Stothard, Reference Stothard2000; available at http://www.bioinformatics.org/sms2/rest_map.html). For determination of PCR-linked random-fragment-length polymorphism (RFLP), ITS1 and ITS2 PCR products from A. erinacei were restricted directly with 2.5 endonuclease units and were incubated 3 h at 37°C. Digests were separated on 2% agarose-TBE gels.
In order to assess the similarity among all sequences of A. erinacei obtained in this study, we analyzed the number of base differences per sequence among all of them using number of differences method of MEGA 5 program version 5.2 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011). Furthermore, we complemented these analyses with other Pulicidae species sequences obtained from GenBank. On the other hand, similarity sequence divergence of cox1 sequences were calculated using the Kimura 2 parameter (K2P) distance model in order to apply the 10X rule (Hebert et al., Reference Hebert, Cywinska, Ball and De Waard2003) and to figure out the threshold level of nucleotide divergence to represent different categories of ‘species’ used by Hebert et al. (Reference Hebert, Cywinska, Ball and De Waard2003). This method was included in MEGA program version 5.2 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).
Phylogenetic trees were inferred using nucleotide data and performed using two methods: Maximum Likelihood (ML) trees were generated using the PHYML package from Guindon & Gascuel (Reference Guindon and Gascuel2003) whereas Bayesian inferences (B) were generated using MrBayes-3.2.6 (Ronquist & Huelsenbeck, Reference Ronquist and Huelsenbeck2003). JMODELTEST (Posada, Reference Posada2008) program was used to determinate the best-fit substitution model for the parasite data (18S, ITS1, ITS2, cox1, and cytb). Models of evolution were chosen for subsequent analyses according to the Akaike Information Criterion (Huelsenbeck & Rannala, Reference Huelsenbeck and Rannala1997; Posada & Buckley, Reference Posada and Buckley2004). For the study of the dataset containing the concatenation of four markers (18S, ITS2, cox1, cytb), analyses based on BI were partitioned by gene and models for individual genes within partitions were those selected by jModeltest. For ML inference, best-fit nucleotide substitution models included general time-reversible model with gamma-distributed rate variation and a proportion of invariable sites, GTR + I + G (ITS2, cox1), transition model with gamma-distributed rate variation, TIM + G (cytb) and general time-reversible model with gamma-distributed rate variation GTR + G (18S and ITS1). Support for the topology was examined using bootstrapping (heuristic option) (Felsenstein, Reference Felsenstein1985) over 1000 replications to assess the relative reliability of clades. The commands used in MrBayes-3.2.6 for BI were nst = 6 with invgamma rates (ITS2 and cox1), nst = 2 with gamma rates (cytb) and nst = 6 with gamma rates (18S and ITS1). For BI, the standard deviation of split frequencies was used to assess if the number of generations completed was sufficient; the chain was sampled every 500 generations and each dataset was run for 10 million generations. Adequacy of sampling and run convergence was assessed using the effective sample size diagnostic in TRACER program version 1.6 (Rambaut & Drummond, Reference Rambaut and Drummond2007). Trees from the first million generations were discarded based on an assessment of convergence. Burn-in was determined empirically by examination of the log likelihood values of the chains. The Bayesian Posterior Probabilities (BPP) was percentage converted.
The phylogenetic analyses, based on18S rRNA, ITS1, ITS2, cox1 and cytb mtDNA sequences were carried out using our sequences and those obtained from GenBank database (Appendix 1). Phylogenetic trees based on 18S rRNA, ITS2, cox1, cytb mtDNA and concatenated (18S, ITS2, cox1, and cytb) sequences were rooted including outgroup species representing members of the Order Mecoptera: Panorpa meridionalis. This choice was based on the combination of morphological and molecular data obtained in former studies which provided compelling evidence for a sister group relationship between Mecoptera and Siphonaptera (Whiting, Reference Whiting2002; Whiting et al., Reference Whiting, Whiting, Hastriter and Dittmar2008). The ITS1 sequence of Panorpa meridionalis or other species of Mecoptera was not available neither by amplification of different individuals nor in any public database. Thus, phylogenetic tree with other Siphonaptera species based on ITS1 sequences were constructed using different outgroup species representing members of Order Diptera: Anopheles moucheti nigerensis and Anopheles moucheti bervoetsi. Thus, ITS1 was discarded for the concatenated dataset. The selection of flea taxa for the concatenated phylogenetic tree was limited to flea species whose 18S, ITS2, cox1, and cytb sequences were available on GenBank database.
Results
Morphological and biometrical results
In total 48 fleas: 13 fleas from two hedgehogs (E. europaeus) and 35 fleas from three hedgehogs (E. europaeus) were collected from Corse and Seville, respectively.
Specific morphological identification done by ourselves was in agreement with that made by our colleagues. Thus, all specimens isolated in this work showed specific morphological characteristics of A. erinacei (fig. 1a–f). Within this species, males of A. erinacei from Corse presented typical morphological characteristics of A. e. erinacei (See material and methods) (fig. 1g), while those from Seville presented typical morphological characteristics of A. e. maura (See material and methods) (fig. 1h) (table S2). Furthermore, total length and total width of the basimere appeared as a significant value to differentiate males from both geographical regions.
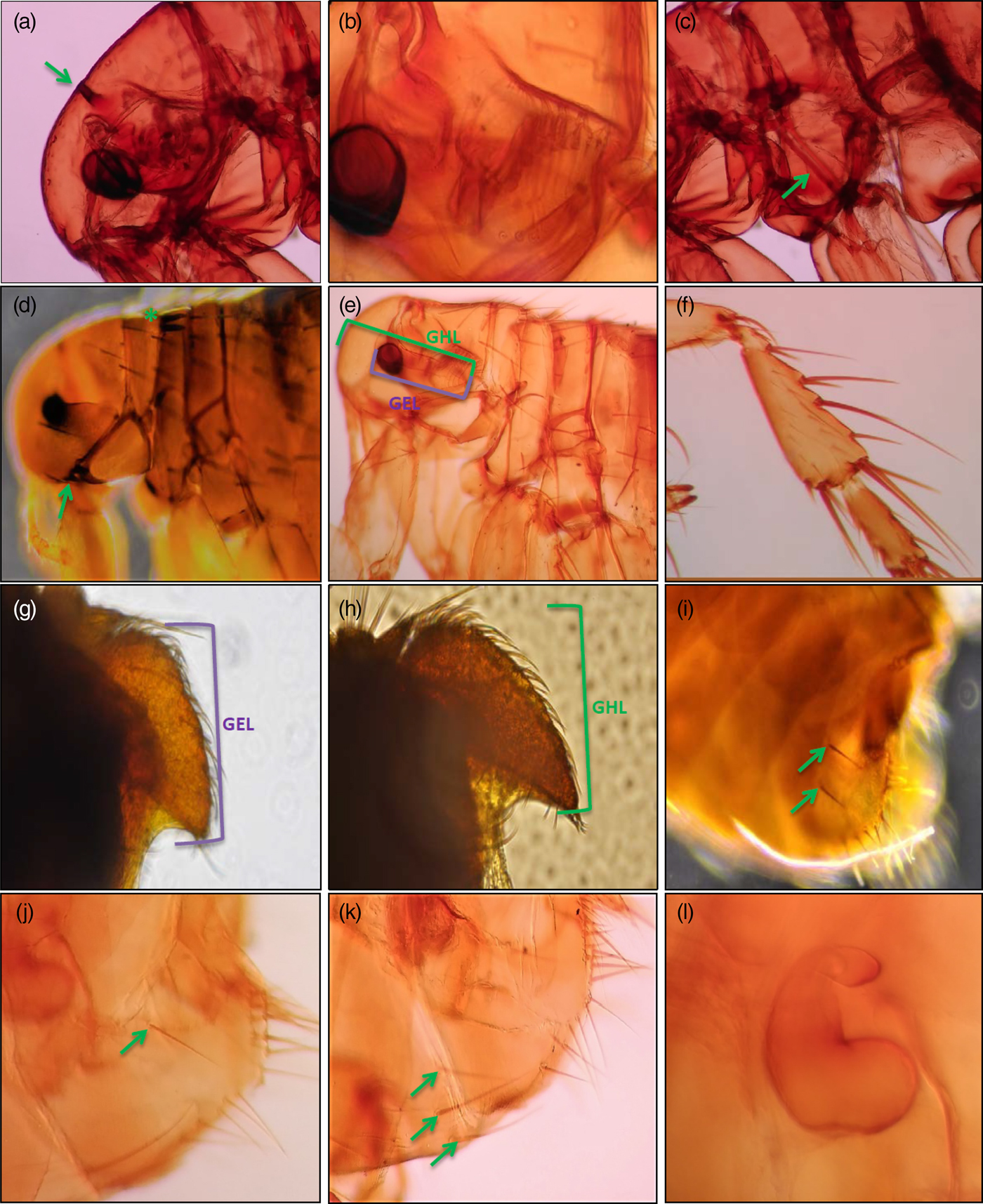
Fig. 1. Morphological specific and subspecific characteristics of Archaeopsylla erinacei and their subspecies: A. e. erinacei (Bouché, Reference Bouché1835) and A. e. maura. (a) Falx of head (arrowed); (b) Asymmetrical antenna with partially welded basal segments; (c) Pleural rod of mesothorax (arrowed) (d) Vestigial genal (arrowed) and pronotal (asterisk) combs; (e) A. erinacei without pronotal comb, GHL: distance from base of spine at tip of genal process to front margin on head, GEL: distance from base of spine on genal process to anterior edge of eye; (f) Hind tibia of A. erinacei; (g) Male basimere of A. e. erinacei; (h) Male basimere of A. e. maura; (i) Female of A. erinacei eighth tergum bearing two lateral bristles (arrowed); (j) Female of A. erinacei eighth tergum bearing only one lateral bristle (arrowed); (k) Female of A. erinacei seventh sternum with three lateral bristles (each side) (arrowed); (l) Spermatheca of A. erinacei.
On the other hand, according to previous morphological descriptions of different authors (See material and methods), we found three different operational taxonomic units (OTUs) of A. erinacei females:
• Population A: A. erinacei females which showed morphological characteristics of A. e. erinacei (See material and methods) (fig. 1i). This population was observed on hedgehogs from Corse and Seville (table S2).
• Population B: A. erinacei females which showed eighth abdominal tergum bearing only one lateral bristle towards the base (fig. 1j) and seventh sternum bore five lateral bristles on the two sides together (two and three bristles each side). This population could not be classified either A. e. erinacei or A. e. maura since it showed ambiguous morphological characteristics. This population was observed on hedgehogs from Corse and Seville (table S2).
• Population C: A. erinacei females which showed eighth abdominal tergum bearing only one lateral bristle towards the base (fig. 1j) and seventh sternum bore six lateral bristles on the two sides together (three bristles each side) (fig. 1k). This population could neither be classified A. e. erinacei nor A. e. maura since it showed ambiguous morphological characteristics. This population was only observed on hedgehogs from Seville (table S2).
Biometrical data (table 2) showed that total width, total length of the head, total width of the head, and the total length of spermatheca (fig. 1l) in females, showed significant values to differentiate females from both geographical regions. Thus, the length of spermatheca was considerably higher in females from Seville than that in females collected from Corse, regardless which OTU they belong.
Mean values of all the morphological traits are showed in table 2. Thus, ANOVA showed several significant differences (P < 0.05) between A. erinaceir (females and males) from Seville and Corse. With respect to females, total width of the female head (HWF) and total length of the spermatheca (EL) showed significant differences, corresponding with the highest values to females from Seville. On the other hand, males showed significant differences (P < 0.05) in total male length (TLM), total length of the male head (HLM), total length of the basimere (BL), total width of the basimere (BW) and total male length of the metathorax (METLM). Thus, males of A. e. maura from Seville presented higher mean of BL and BW than those from Corse and, by contrast, males from Corse presented higher mean of TLM, HLM and METLM than those from Seville (Spain).
Molecular results
Partial 18S rRNA gene analysis
Partial 18S rRNA gene sequences of different populations of A. erinacei were 1160 base pairs (bp) in length (table 1). No differences were observed between partial 18S rRNA gene sequences from both geographical origins. Partial 18S gene phylogenetic tree showed species belonging to Pulicidae family clustered together, with high bootstrap and Bayesian Posterior Probabilities (BPP) values, but phylogenetically distant from Stenoponiidae, Ctenophthamidae, and Ceratophyllidae (tree not shown). Nevertheless, this tree was unable to differentiate at species and subspecies level.
ITS1 and ITS2 analysis
The length of the ITS1 sequences of A. erinacei ranged from 949–950 (Seville) to 951 (Corse) (table 1). On the other hand, ITS2 sequence length ranged from 360 (Corse) to 361 (Seville). This length difference was also observed in clones from individuals from two different geographical origins and was due to the existence of one extra basis pair in position 258 in the ITS2 sequence of the individuals from Seville.
ITS2 intra-individual similarity studied in seven clones of one individual of A. erinacei from Corse ranged from 99.4 to 100%, whereas this value ranged from 99.2 to 100% when eight clones of one individual of A. erinacei from Seville were compared. Specimens obtained from the same geographical area showed the same ITS2 sequence (Intra-population similarity = 100%), indistinctly if they belong to different morphological populations (females). Unlike this value, when the ITS2 sequences of individuals from both geographical origins (Corse and Seville) were compared, the similarity observed was 96.9% (Intra-specific similarity = 96.9%).
ITS1 sequences of specimens from the same geographical origin were identical (Intra-population similarity = 100%). On the other hand, when the ITS1 sequences from both geographical origins were compared, the similarity observed was 99.1% (Intra-specific similarity = 99.1%).
Based on ITS1 and ITS2 sequences, restriction mapping identified endonucleases delineating the two different geographical areas (Corse and Seville) (fig. 2). Thus, EcoRV, HaeIII, and PhoI presented one restriction site in ITS1 sequences of A. e. erinacei (male) from Seville but none in A. e. maura (male) from Corse (fig. 2). Restriction mapping for ITS2 sequences showed AseI, MseI (Position 78) and VspI presented one restriction site in A. e. erinacei (male) from Corse but none in A. e. maura (male) from Seville, whereas, AsuII, BbuI, DraI, NIaIII, PsiI, MseI (Position 179) and SphI presented one restriction site in ITS2 sequences of A. e. maura from Seville but none in A. e. erinacei from Corse (fig. 2). The endonuclease HaeIII was chosen for the use in the PCR-linked RFLP analysis of ITS1. As predicted by the sequence data, restriction of ITS1 PCR products of A. erinacei from two geographical origins with HaeIII produced two restriction fragments (194 and 755 bp) for individuals from Seville and an undigested product (951 bp) for individuals from Corse (fig. 3).

Fig. 2. (a) Schematic representation of restriction maps of the ITS1 sequence of A. e. maura collected from Seville. (b) Schematic representation of restriction maps of the ITS2 sequence of A. e. maura collected from Seville. (c) Schematic representation of restriction maps of the ITS2 sequence of A. e. erinacei collected from Corse.

Fig. 3. PCR-RFLP analysis of the ITS1 of A. erinacei collected from different geographical origins using HaeIII endonuclease. M = DNA Molecular Weight Marker IX (72–1353 bp); Line 1 = A. e. erinacei from Seville; Line 2 = A. e. maura from Corse.
The phylogenetic tree inferred from ITS2 sequences of A. erinacei and other ITS2 sequences retrieved from GenBank (see Appendix 1) showed all Pulicidae species clustered together with high bootstrap and BPP values and phylogenetically close to Stenoponiidae family (fig. S1). Within Pulicidae clade, A. erinacei specimens comprised a well-supported subclade phylogenetically related to the remaining Pulicidae species. This subclade showed individuals separated according to geographical origin with high bootstrap and BPP values, indistinctly these individuals belong to different morphological populations (fig. S1).
ITS1 phylogenetic tree revealed a subclade clustering all A. erinacei specimens related to Ctenocephalides within Pulicidae family clade. Furthermore, likewise in ITS2 phylogenetic tree, A. erinacei individuals clustered separated according to geographical origin with high bootstrap and BPP values (fig. S2).
Partial cox1 and cytb mtDNA gene analysis
The partial cox1 mtDNA gene sequences of A. erinacei from the two geographical areas were 658 bp in length (table 1). Intra-population similarity observed ranged from 99.8 to 100% in both geographical origins, while intra-specific similarity ranged from 97.7 to 98.1% (table 3). Furthermore, the conspecific divergence ranged from 0 to 0.2. If we consider that the average of conspecific divergence was 0.09, we can apply the 10X rule; thus, the threshold level of nucleotide divergence between two Archaeopsylla species would be 0.9%. Nevertheless, any value of conspecific divergence among all individuals analyzed in this study overcame this threshold.
Table 3. Intra-population and intra-specific similarity observed among all the partial cox1 mtDNA gene sequences of Archaeopsylla erinacei from different geographical areas obtained in this work and other Pulicidae species from GenBank database.

Values are given in percentages. (AE = A. erinacei).
On the other hand, the length of the partial cytb mtDNA gene sequences of A. erinacei from Corse and Seville was 374 bp (table 1). Intra-population similarity of A. erinacei specimens from Seville ranged from 98.1 to 100%, while this value was 100% for specimens collected from Corse. Intra-specific similarity ranged from 98.1 to 98.9% (table 4). Furthermore, inter-specific cytb similarity observed between others congeneric species belonging to Pulicidae family showed quite lowest percentage values than those observed between A. erinacei specimens from the two different geographical origins analyzed in this work (table 4).
Table 4. Intra-population and intra-specific similarity observed among all the partial cytb mtDNA gene sequences of Archaeopsylla erinacei from different geographical areas obtained in this work and other Pulicidae species from GenBank database.

Values are given in percentages. (AE = A. erinacei).
Phylogenetic tree topology of both mitochondrial markers revealed a highly supported clade clustering all Pulicidae species (figs S3 and S4). In addition, A. erinacei individuals from Seville clustered together with high bootstrap and BPP values and separated from A. erinacei specimens collected from Corse indistinctly if these individuals belong to different morphological populations (figs S3 and S4). Particularly, in cox1 phylogenetic tree, Ctenocephalides species appeared clustering near to Archaeopsylla with high bootstrap and BPP values (96/82), whereas in cytb phylogenetic tree, Ctenocephalides species and the others Pulicidae species clustered in polytomy in relation to Archaeopsylla.
The concatenated dataset of the partial 18S gene, ITS2, partial cytb, and cox1 gene sequences included 2558 aligned sites and 30 taxa, including outgroups. Phylogenetic analyses of the concatenated dataset yielded a tree with branches strongly supported (fig. 4). The analysis based on the concatenated dataset is concordant with all trees constructed on the basis of the single markers. Thus, all species belonging to Pulicidae family clustered together in two main subclades with high bootstrap and BPP support. The first one clustered all Ctenocephalides species, while in the second one all Archaeopsylla species clustered separated according to two different geographical origins: Corse and Seville (fig. 4).

Fig. 4. Phylogenetic tree of Archaeopsylla erinacei from different geographical origins (see Table 1) based on concatenated partial 18S ribosomal RNA gene, Internal Transcribed Spacer 2 (ITS2) partial cytochrome c-oxidase 1 (cox1) and cytochrome b (cytb) gene of mitochondrial DNA inferred using the Bayesian (B) method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown on the branches. The Bayesian Posterior Probabilities (BPP) are percentage converted.
Discussion
It has been widely reported the idea that majority of characters used for flea species and subspecies diagnoses are based on the shape and structure of their extraordinarily complex genitalia, or the presence and distribution of setae and spines (Traub & Starcke, Reference Traub and Starcke1980; Dunnet & Mardon, Reference Dunnet, Mardon, Naumann, Lawrence, Nielsen, Spradberry, Taylor, Whitten and Littlejohn1991). While these characters are adequate for species diagnoses, they are mostly autapomorphic at the species and subspecies level and of limited utility for phylogenetic reconstruction. Thus, Siphonaptera appears to have many instances of parallel reductions and modifications, probably associated with multiple invasions of similar hosts, which may obscure homology. In addition, from a phylogenetic standpoint, Siphonaptera has remained as the most neglected of the holometabolous insect orders (Whiting et al., Reference Whiting, Whiting, Hastriter and Dittmar2008).
The present work represents the first study that provides morphological, biometrical, molecular and phylogenetic comparative data of A. erinacei and their subspecies: A. e. erinacei and A. e. maura, in order to assess taxonomic and phylogenetic relationships between both subspecies and to shed light on the systematics of A. erinacei, representing a new tool to elucidate identification within the genus.
From a morphological standpoint, Jordan & Rothschild (1953) were the first authors who provided some morphological features in order to identify and discriminate between both subspecies. They based the male morphological identification on the length of basimere, whereas female morphological subspecies discrimination was based on the presence of one or two lateral bristles in eighth abdominal tergum and the presence of four or five lateral bristles in seventh abdominal sternum on the two sides together. Beaucournu & Launay (Reference Beaucournu and Launay1990) accepted these morphological criteria in order to discriminate both subspecies, excluding the setae number observed in the seventh abdominal sternum. Nevertheless, these authors pointed out the high taxonomic similarity between these two subspecies and they observed that only male specimens could be identified easily each other. Our results reinforce the idea of the use of the length of basimere as a useful morphological criterion in order to discriminate between males of A. e. maura and A. e. erinacei. Thus, based on these criteria we conclude that males collected from Corse belong to A. e. erinacei, while male specimens collected from Seville belong to A. e. maura.
Unlike male individuals, our results showed that previous criteria used for morphological differentiation in females of A. erinacei were not useful to discriminate between both subspecies. Thus, we observed different morphological populations of females showing overlapped morphological characters that not corresponded with any previous subspecific morphological characterization cited by different authors. Furthermore, a geographical pattern of distribution was not observed in female specimens, appearing A. e. erinacei (population A) and population B in both geographical areas. With these results, two different hypotheses could be suggested. The first one would consider that A. e. erinacei occurs in both geographical areas and the appearance of population B and C just mean morphological variants belonging to a polymorphic taxon. The other one could be considering that the morphological classification of females does not support the male one, therefore, it could be suggested to discriminate between both subspecies based exclusively on the morphological characteristics of the male specimens unless new discriminative morphological characters were revealed for female subspecific classification. In this sense, we observed, by the first time, that the total length of the spermatheca could be a useful criterion in order to discriminate between both females’ subspecies since this criterion display a geographical pattern of distribution corroborated by molecular and phylogenetic data. Thus, we could conclude that individuals from Seville showing a total length of spermatheca higher than 120 µm corresponded with A. e. maura while those from Corse showing a total length of spermatheca lower than 120 µm corresponded with A. e. erinacei. Furthermore, length of spermatheca appeared as a significant value calculated by ANOVA test to differentiate both subspecies. Nevertheless, we assumed that future research based on the study of populations of A. erinacei from different geographical areas could consolidate and support the taxonomic status of this species.
The analysis of external morphological characters presents some weaknesses when are used as the unique criterion to distinguish female specimens of this species. Thus, the use of molecular biology is considered as an essential tool in order to clarify morphological data.
These facts, lead us to suggest that A. erinacei subspecies might have been morphologically misidentified for many years in Mediterranean area. This observation could be the consequence of a wrong identification practice of females based on morphological differences of male specimens or the GO as a valid criterion for the identification between both subspecies. Lewis (Reference Lewis1967) and Beaucournu & Launay (Reference Beaucournu and Launay1990) argued that certain flea subspecies admitted by some authors could just be a morphological variant belonging to a polymorphic taxon. This fact is corroborated by phylogenetic analyses in our study, in which we did not find correspondence between female morphological differences analyzed and the 18S, ITS1, ITS2, cox1, and cytb sequences.
According to ITS's analyses, ITS2 sequences of both subspecies were markedly shorter than ITS1 sequences. Vobis et al. (Reference Vobis, D'Haese, Mehlhorn, Mencke, Blagburn, Bond, Denholm, Dryden, Payne, Rust, Schroeder, Vaughn and Bledsoe2004) and Zurita et al. (Reference Zurita, Callejón, De Rojas, Gómez-López and Cutillas2015, Reference Zurita, Callejón, De Rojas, Halajian and Cutillas2016, Reference Zurita, Callejón, De Rojas and Cutillas2018) have previously reported this fact in other species of fleas such as C. felis, Stenoponia tripectinata tripectinata, C. canis, N. barbarus, and N. fasciatus.
Both markers (ITS1 and ITS2) did not show sequence differences among individuals from the same geographical area regardless they belong to different morphological populations (females). Nevertheless, they showed different percentage of similarity ranged from 96.9% (ITS2) to 99.1% (ITS1) between specimens from two geographical regions each other. Thus, these nuclear markers were useful to differentiate A. erinacei from Seville and Corse. Marrugal et al. (Reference Marrugal, Callejón, de Rojas, Halajian and Cutillas2013) reported similar values of similarity and Zurita et al. (Reference Zurita, Callejón, De Rojas, Halajian and Cutillas2016), who reported an inter-specific similarity ranged from 91.8 to 96% between ITS sequences of C. felis and C. canis isolated from dogs from different geographical areas. These geographical signals in fleas have previously been reported by Luchetti et al. (Reference Luchetti, Trentini, Pampiglone, Fiorawanti and Mantovani2007), who noticed the presence of two genotypic groups (Pacific and Atlantic) based on the analysis of ITS2 sequences of Tunga penetrans from Ecuador, Brazil and different geographical areas of Africa. In addition, several specific recognition sites for endonucleases were detected in ITS1 and ITS2 sequences in order to differentiate two geographical lineages. Thus, EcoRV, HaeIII, PhoI, AseI, VspI, AsuII, BbuI, DraI, NIaIII, MseI, PsiI, and SphI sites have diagnostic value for specific determination of subspecific discrimination in A. erinacei.
The partial cox1 and cytb mtDNA gene sequences showed the same geographical pattern than ITS sequences analyses (tables 3 and 4) regardless which morphological population they belong to. On the other hand, cox1, cytb, and concatenated phylogenetic trees reinforce the idea of the existence of two geographical genetic lineages in A. erinacei (Iberian Peninsula and Corse Island). Furthermore, cox1 phylogenetic tree showed specimens belong to Ctenocephalides and Archaeopsylla genera clustered together. Zhu et al. (Reference Zhu, Hastriter, Whiting and Dittmar2015) who included both genera in Archaeopsyllini subfamily reported this close phylogenetic relation between Ctenocephalides and Archaeopsylla genera.
Previous studies showed that fleas have a high level of intraspecific genetic variation (Dittmar & Whiting, Reference Dittmar and Whiting2003; Brinkerhoff et al., Reference Brinkerhoff, Martin, Jones and Collinge2011). Furthermore, it has been suggested that host specificity may influence the level of intraspecific genetic divergences since more generalist parasite species will show a higher level of intraspecific genetic variation enabling them to infest a broader host range (Van der Mescht et al., Reference Van der Mescht, Matthee and Matthee2015). DNA barcoding studies on insects and invertebrates have shown maximum intra-specific variation ranging from 3 to 3.9% (Carew et al., Reference Carew, Pettigrove, Cox and Hoffmann2007), out of which are markedly higher when specimens of study come from distant geographical regions, especially islands or archipelagos. In this way, Lawrence et al. (Reference Lawrence, Brown, Peters, Spielman, Morin-Adeline and Slapeta2014), Zurita et al. (Reference Zurita, Callejón, De Rojas, Gómez-López and Cutillas2015) and Zurita et al. (Reference Zurita, Callejón, De Rojas and Cutillas2018) found a high degree of intra-specific variation in some flea species when populations from islands and mainland were compared, suggesting the existence of different geographical lineages, which could have arisen due to the existence of geographical barriers.
The cox1 similarity values observed between both geographical genetic lineages (97.7–98.1%) in A. erinacei were similar with those observed among different flea species such as C. felis and C. canis (97.7) (table 3). This fact could suggest that individuals from Spain and Corse could be treated as different species. Nevertheless, based on K2P analysis and 10X rule reported by Hebert et al. (Reference Hebert, Cywinska, Ball and De Waard2003) we cannot assume that both geographical genetic lineages correspond with two different species within Archaeopsylla genus.
Our results are in agreement with Losos & Ricklefs (Reference Losos and Ricklefs2009) who suggested that detailed population-level studies can chart the course of evolution over short time periods. This approach can be broadened to incorporate intra-specific level studies with a geographically explicit sampling of individuals for the reconstruction of gene genealogies to reveal the extent to which natural selection or alternative mechanisms may explain evolutionary change. In this sense, island radiations are ideal systems for such an approach, because it is frequently apparent that the arena within which inter-specific diversification has occurred is similar to the arena within which intra-specific diversification is occurring (Ricklefs & Bermingham, Reference Ricklefs and Bermingham2001).
In conclusion, the present study provides for the first time, comparative morphological, biometrical, and molecular data of A. erinacei and their subspecies: A. e. erinacei and A. e. maura. On the basis of morphological results, we conclude that the number of bristles bearing in eighth abdominal tergum and seventh abdominal sternum of female specimens are not valid criteria as diagnostic characters in order to differentiate A. e. erinacei and A. e. maura. However, the total length of the spermatheca in females and the different length of basimere in males should be taking into account as characters of reference in order to discriminate between both subspecies.
On the other hand, based on phylogenetic and molecular comparative study of two nuclear markers (ITS1 and ITS2), two mitochondrial markers (cox1 and cytb) and concatenated sequences, we reported the existence of two geographical genetic lineages in A. erinacei corresponding with two different subspecies (A. e. erinacei and A. e. maura), that could be discriminated by PCR-linked RFLP.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485317001274.
Acknowledgement
The present work was supported by a grant of the V Plan Propio de Investigación of the University of Seville, Spain. The authors thank Dr Carlos Feliu (University of Barcelona) for providing samples from Corse (France).











