Published online by Cambridge University Press: 15 July 2005
Population genetics of multi-host pathogens offers great potential for the understanding of their complex epidemiology but care must be taken to ensure that the sampling procedure does not bias estimates of population indices. The transfer of material to laboratory passage, in particular, runs the risk of bottlenecking and imposing non-random host-induced selection pressures according to the hosts used in passage. We present a novel technique allowing single-locus microsatellite genotyping of the naturally sampled larval stages, enabling unbiased population genetic studies of the multi-host zoonotic parasite Schistosoma japonicum. The utility of these larval genotyping methods for molecular epidemiological studies are illustrated in results from 3 separate data sets. In the first data set, potential loss of alleles based on the definitive host species used for laboratory maintenance was identified by comparing adult worm populations derived from mice and rabbits infected with cercarial populations originating from the same set of snails. In the second data set, bottlenecking was demonstrated by the loss of alleles in adult worms derived within a single generation of laboratory maintenance compared to their parent field-collected cercarial samples. In the final data set, comparison of miracidia and adult worms recovered from naturally infected animals demonstrated that larval analyses can provide stage-specific epidemiological information and that population genetics of schistosomes can be well described by analysis of larval stages. Our results thus advocate the use of natural life-cycle stages to obtain an accurate and ethical representation of the population genetic structure of S. japonicum and other multi-host pathogens.
Many important human, domestic and wildlife diseases are caused by parasites able to infect multiple host species (Taylor, Latham and Woolhouse, 2001), including, for example, in excess of 60% of human pathogen species (termed zoonoses) and 77% of livestock pathogens (Haydon et al. 2002). Whilst population geneticists and biologists have been very successful in developing a formal understanding of the dynamics and evolution of single host pathogens, the understanding of the more complex population biology of multi-host pathogens is severely limited and has been declared as one of the major emerging challenges of the biomedical sciences in the 21st century (Woolhouse, Taylor and Haydon, 2001). Generalizations between single-host and multi-host pathogens often cannot be made because the machinery required for infection, exploitation, and transmission, as well as the selective host-induced pressures acting on parasites, are likely to vary from one host to another (Gandon, 2004). Moreover, the study of population genetics in multi-host pathogens becomes increasingly difficult because it can encompass both polymorphism (differences among organisms of the same species) as well as divergence (differences that accumulate between species) and, as a result, the parasites are often referred to as variants on the basis of genotypes and phenotypes within either the same host or different host species (Taylor et al. 2001).
The zoonotic Schistosoma japonicum may provide an important illustration. S. japonicum, a major human pathogen, is endemic within The Philippines, China and Indonesia. In China, for example, it is estimated that approximately 865000 people are infected and 40 million are at risk (Chen and Feng, 1999). A high equilibrium prevalence of infection has persisted despite implementations of human chemotherapy in the 1960's, thereby emphasizing the need to understand and quantify the ecology and transmission of this parasite (WHO, 1980). Like all schistosomes, S. japonicum is a digenean trematode with an indirect life-cycle involving sexual reproduction in a mammalian host and an asexual phase in a molluscan host. Transmission between hosts occurs via free-swimming larval stages, miracidia (infective to the mollusc) and cercariae (infective to the mammal). S. japonicum is unique, however, amongst the human schistosomes in that over 40 species of wild and domesticated non-human mammals are suspected to act as reservoir hosts for the parasite (Dumag et al. 1981; He, Salafsky and Ramaswamy, 2001). The potential role that domesticated animals play in transmission of the parasite to each other and the human population is now being acknowledged, and indeed the Tropical Research Unit of the WHO have recently prioritised development of a vaccine for livestock S. japonicum as a way of controlling human infection. However, there exists a serious lack of epidemiological data on S. japonicum infection in animals, and hence their actual role in transmission, upon which any appropriate and sustainable surveillance, prevention and control strategies must be based, remains unknown (Hagan and Gryseels, 1994).
Molecular epidemiological studies of S. japonicum present a valuable opportunity to gain much needed insight into the transmission and epidemiology of this complex parasite. A key problem in studies of schistosome epidemiology, however, is that adult worms are not available for direct sampling from human and valuable domestic livestock because of their location in the blood vessels of the mesenteric system surrounding the intestine. Direct characterization of adult schistosome worms of natural non-human hosts provides an alternative approach (e.g. S. mansoni population genetic surveys have been carried out by passaging wild rats in Guadeloupe (Theron et al. 2004)), but care must be taken in interpretation of such results since schistosome transmission may be quite different in humans and indeed within other definitive hosts. Previous work on S. japonicum has thus been limited to the study of adult worms generated by a single generation (He et al. 1994; Bogh et al. 1999; Chilton et al. 1999; Gasser et al. 1996) or multiple generations (Bowles et al. 1993; Merelender et al. 1987) of laboratory passage. This involves hatching eggs from the faeces of infected humans or animals and infecting laboratory snails with miracidia and/or exposing laboratory mammals to cercariae to allow development of adult worms. In the majority of cases only a single species of laboratory mammal is used for passage (e.g. Bogh et al. 1999; Merelender et al. 1987 but see He et al. 1994). In addition to the ethical and practical implications, in terms of the necessity for large numbers of laboratory mammal infections, such laboratory passage may be predicted to result in genetic bottlenecking of the parasite and impose selection pressures not encountered in natural infections, thereby resulting in parasite isolates unrepresentative of the genetic variation of the original population. A particular concern for S. japonicum, as indeed for all zoonotic parasites, is the further biasing due to species-specific host-imposed selection by the laboratory mammals used for isolate collection, where one may predict, for example, differential success of rodent-biased genotypes within laboratory rodent passage since rodents represent one natural reservoir (Dumag et al. 1981; He et al. 2001). Another important concern for population genetic studies of S. japonicum by using infections to laboratory animals is with respect to the widely observed trickle infections (Hurst, 2000). In endemic areas, humans and other definitive hosts are likely to be continuously exposed to repeated low-level infections which lead to accumulation of parasite genomes. Since these infections are very difficult to emulate experimentally in laboratory animals such as mice and hamsters because of their short life-span (Bergquist, 1995), studies have been based on only a single infection of parasites given once (Chilton et al. 1999; Stohler, Curtis and Minchella, 2004; Shrivastava et al. 2005, but see Hurst, 2000), which can further bias the results because all natural infections might not be sampled.
We report here novel methodology for the storage and subsequent polymerase chain reaction (PCR) amplification and microsatellite analysis of free-living larval stages of S. japonicum that can be directly sampled from natural populations. We demonstrate the utility of these assays by presenting 3 data sets which suggest, also for the first time, that the method of isolate collection for S. japonicum has the potential to seriously bias results of population genetic analysis.
Three independent data sets were used here to assess the utility of the miracidial and cercarial assays on natural schistosome populations and the potential disadvantages of standard isolate collection methods. The parasite samples from within Mainland China were collected from endemic regions of Anhui province of Southeast China, which is one of the 5 provinces in the swamp and lake regions of China where schistosomiasis is highly prevalent (Chen, Jiang and Zhao, 2001; Yang et al. 2004). Similarly, S. japonicum samples from The Philippines were collected from highly endemic and swampy regions of Samar and Leyte province situated within Southeast Philippines (Fernandez, Petilla and Banez, 1982).
Infected Oncomelania hupensis snails were collected within a 10 km2 sampling area from Guichi village of Anhui province in South China and transported to the laboratory (Zhejiang) (Shrivastava et al. 2005). Fifty infected snails were induced to shed and the resulting cercariae were pooled to percutaneously infect 2 rabbits at a dose of 1000 cercariae per animal and 20 laboratory mice at a dose of 200 cercariae per animal according to standard protocols (Johansen, Christensen and Nansen, 1997). After 45 days, adult schistosomes were collected from the mesenteric veins by perfusion. Worms from the 2 rabbits and from 5 randomly selected mice were pooled separately to create ‘rabbit-passaged’ and ‘mouse-passaged’ isolates. Fifty randomly chosen individual S. japonicum adult worms (equal mix of male and female worms) from these two sample isolates were selected for this study.
Ten patent O. hupensis snails were collected from Barangay Cabarsan Guti, town-Palo, and province-Leyte in the Philippines and transported to the Natural History Museum, London, UK, under guidelines issued by the Department for Environment, Food and Rural Affairs (DEFRA)-UK. The snails were induced to shed cercariae and 7 snails that were shedding more than 300 cercariae (following shedding for 12–16 h) were chosen for this analysis. A total of 150 cercariae from each individual snail were used to infect a single mouse, and hence there were 7 mice individually infected with cercariae from single snails. In addition, 120 cercariae per snail were individually stored for direct microsatellite analysis.
Parasite samples from domesticated goats were collected from Shanlian village, Dangtu County, located in the island within the lower region of the Yangtze River within Anhui Province, China. Two goats naturally infected with S. japonicum (identified by the presence of schistosome eggs in their faeces) were used to obtain miracidia and adult worms. Thus 40 randomly chosen individual S. japonicum adult worms (equal mix of male and female), and 240 randomly selected miracidia hatched from faeces of the same two goats (20 miracidia/locus/goat) were analysed.
Similarly, parasite samples from wild rats were collected from the Barangay of Hinucagan, Municipality of Gandara, and island of Samar in The Philippines. Two rats infected with S. japonicum were identified by hatching of miracidia from faecal samples and 80 worms were recovered from the mesenteric vessels and 120 miracidia successfully hatched from the faeces. All samples were subject to microsatellite analysis.
Adult worms. Adult worms were recovered from the mesenteric veins of definitive hosts by hepatic perfusion using 0·15 mM sodium chloride and 25 mM sodium citrate (Duvall and DeWitt, 1967). Worms were washed extensively in physiological saline and stored in 100% ethanol until further use (Bogh et al. 1999).
Miracidia were hatched from faecal samples and cercariae induced to shed from snails in autoclaved deionized water according to standard protocols (Bogh et al. 1999; Stohler et al. 2004). In order to minimize the presence of contaminants, 3 sequential washings in autoclaved deionized water were conducted by repeated transfer of individual miracidia or cercariae to successive Petri dishes containing autoclaved deionized water using a glass pipette under the binocular microscope. Following washing, individual larvae were collected in sterile 0·2 ml PCR tubes using a minimum quantity of autoclaved deionized water and immediately snap frozen to –70 °C, where they were stored for up to 3 months before use. These conditions had been previously standardized following experiments to validate various storage conditions (Table 1).

Genomic DNA was extracted from individual schistosomes using a phenol–chloroform procedure as described by (Barker and Bundy, 1999) and purified using GeneClean Kit II (Bio101-UK).
The tubes containing single miracidia or single cercariae were vacuum dried (Speed Vac-Thermo Savant-UK) to reduce the final volume to 5 μl. Two nanograms of Proteinase K (Sigma Aldrich-UK) was added to each tube, and incubated at 55 °C for 1 h. After the incubation, Proteinase K was heat-inactivated by placing the samples at 98 °C for 15 min. Immediately after the heat treatment, the sample tubes were placed over ice for at least 10 min to rapidly cool the samples and then stored at −40 °C for 1 h. The total volume of the genomic DNA was then transferred to a fresh PCR tube for conducting PCR amplification.
All of the 11 previously isolated and characterized S. japonicum microsatellite markers (Shrivastava et al. 2003) showed successful amplifications on single larvae. Six of these loci, namely-2AAA, MF1, M5A, RRPS, J5 and MPA, were used in this study using the published annealing and magnesium conditions. For each locus, the 5′ end of the forward primer was fluorescently labelled using 6-FAM dye (MWG-UK). Polymerase Chain Reactions (PCR) amplifications were performed on a PTC-200 Thermal Cycler (MJ Research-UK). Amplifications were performed in 40 μl reactions with 50 nanograms of adult worm DNA or 7 μl of miracidial genomic DNA, 5 pmoles of each primer, 1 unit of Taq polymerase, 4 μl of reaction buffer (10 mM Tris-HCl pH 9·0, 50 mM KCl, 0·1% Triton®X-100; Promega-UK), 0·4 mM dNTP mix (Sigma Aldrich-UK) and 1·5–2·5 mM MgCl2 (Promega-UK). Thermal cycling was conducted under the following conditions: 5 minutes at 94 °C, followed by 35 cycles for adult worms or 50 cycles for larval stages, of 30 sec at 94 °C, 1 min at locus-specific temperature, 1 min at 72 °C, with a final extension at 72 °C for 7 min. Products were diluted in N, N′-dimethyl formamide with GeneScan ®-500[ROX] (for PCR products of size range <500 base pairs) or GeneScan ®-2500[ROX] (for PCR products of size range >500 base pairs) size standard (both ABI-UK) and electrophoresed using an ABI 377 (for PCR products of size range <500 basepairs) or ABI 3100 (for PCR products of size range >500 base pairs). Allele sizes were assigned using GENESCAN software v 3.7 (PE Applied Biosystems-UK) and Genotyper v 3.7 (PE Applied Biosystems-UK). Due to their small size (approximately 0·22 mm by 0·08 mm); each larval sample produced sufficient DNA for only a single PCR reaction.
For each of the 3 data sets, Genetic data analysis (GDA) version 1.1 (Lewis and Zaykin, 2001) was used to calculate the number of alleles, number of polymorphic loci and private alleles (alleles exclusive to any subpopulation).
Arlequin 2.0 (Schneider, Roesli and Excoffier, 2000) was used to perform Hardy-Weinberg equilibrium tests of the hypothesis that observed diploid genotypes are the product of random union of gametes (i.e. the population is randomly mating). To detect significant departure from this equilibrium, a modified version of the Markov-chain random walk algorithm described by Guo and Thompson (1992) was used, a test analogous to Fisher's exact test using contingency table of observed allele frequencies and the number of alleles through 100000 permutations. Because the gametic phases of our loci are not known, this test was only possible for each locus separately. Although this approach does not allow us to estimate association between alleles within individual parasites, previous work with S. japonicum has demonstrated minimal linkage disequilibrium between these loci within individuals (Shrivastava et al. 2005).
As a measure of genetic distance between populations using microsatellite loci, Fst calculations were performed in Arlequin 2.0 (Schneider et al. 2000), which counts the number of different alleles at each locus and between samples as well as takes into account differential population sizes. Unweighted Pair Group Method with Arithmetic Mean (UPGMA) clustering was also performed on pair-wise population co-ancestry genetic distance (calculated in Arlequin 2.0) data using Molecular Evolutionary Genetic analysis (MEGA) version 2.1 (Kumar et al. 2001) that employs a sequential clustering algorithm, in which local topological relationships are identified in order of similarity and a phylogenetic tree is built in a stepwise manner. UPGMA analysis thus demonstrates visually the similarity of individuals or populations, with the length of branches showing the genetic distances separating clusters.
In addition to developing a novel protocol for storage and genotyping of single larval stages (Table 1), using these assays we have been able to perform successful PCR amplification and microsatellite genotyping of individual miracidia and cercariae of S. japonicum. Out of the 11 microsatellite loci amplifiable on single S. japonicum larvae, 6 were selected for this study based on their polymorphic content and absence of null alleles. Allele identifications established using these assays were reliable and straight-forward making these very useful in characterizing both naturally collected and laboratory generated parasite material.
As shown in Table 2, the total number of alleles detected by using both mice and rabbits was high indicating suitability of microsatellite markers for the present study. However, despite the fact that both rabbits and mice were exposed to the cercariae from the same origin, there were alleles detected in adult worms recovered from rabbits which were not detected in adult worms recovered from mice and vice versa. This was true for each of the 6 microsatellite loci studied. Most of these private alleles were rare alleles, i.e. present at a frequency of less than 0·05 within the populations. UPGMA phenogram (Fig. 1) using co-ancestry identity suggested infra-populations from single animals clustered more closely with the infra-populations from the same definitive host species than with those derived from the other definitive host. These results were supported by the Fst value of 0·01–0·04 (95% confidence limits) which revealed low yet significant distance between the samples collected from rabbits and those from mice.


Fig. 1. UPGMA phenogram depicting co-ancestry identity between Schistosoma japonicum adult worms passaged from mice and rabbits. Each population represents 20 worms from an individual definitive host.
Of the 7 mice infected with S. japonicum cercariae described above, only 3 produced patent multiple sex infections and the resultant miracidia. As shown in Table 3, a larger number of private alleles were present in the cercarial samples, suggesting a significant loss of alleles within a single laboratory passage. No definite pattern was observed regarding heterozygosity levels (Table 4). The decrease in heterozygosity (observed heterozygosity compared to expected heterozygosity) within 1 laboratory passage was not significantly different from the larval stages (cercariae) collected directly from the snails.


UPGMA (Fig. 2a, b and c) for the 3 mice that produced miracidia revealed clustering separating cercariae from adult worms and miracidia. This result was consistent with Fst estimates of 0·032–0·246 (95% confidence limits) between pooled cercariae and adult worms and Fst estimates of 0·026–0·173 (95% confidence limits) between pooled cercariae and miracidia within 1 laboratory passage. These values revealed significant distance between the natural samples and samples recovered following laboratory passage.
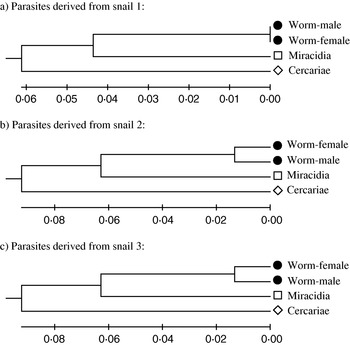
Fig. 2. UPGMA phenogram depicting co-ancestry identity between cercariae and adult worms plus miracidial stages of Schistosoma japonicum obtained from passaging through laboratory mice. The corresponding snails had been collected from Cabarasan Guti, Palo-Leyte.
As shown in Table 5, the microsatellite markers used in this study were highly polymorphic in both miracidia and adult worms. Differing private alleles were identified in both adult worms and miracidia. In other words, there were some alleles present in adult worms that were never observed in miracidia and vice versa. However, on average, the frequency of private alleles was higher in miracidia than in adult worms. The low levels of Fst estimates of 0·03–0·08 (95% confidence limits) and UPGMA phenogram (Fig. 3) suggested that miracidia from the 2 goats may be more similar to each other than to the adult worms from the same infected animals.

Fig. 3. UPGMA phenogram depicting co-ancestry identity between adult and miracidial stages of Schistosoma japonicum obtained from domesticated goats collected form Anhui Province of China.

As shown in Table 6, microsatellite markers were again highly polymorphic in both miracidia and adult worms. Notably, private alleles were only present in miracidia, suggesting that miracidia had all the alleles present in the adult worm population and some extra alleles. These results were supported by the significant yet very low Fst value of 0·005–0·001 (95% confidence limits) between adult worms and miracidia and UPGMA phenogram (Fig. 4) which indicates minimal clustering between adult worms and miracidia sampled from wild rats suggesting suitability of miracidia for studying adult parasite genotype within definitive host species.

Fig. 4. UPGMA phenogram depicting co-ancestry identity between adult and miracidial stages of Schistosoma japonicum obtained from wild rats collected from Barangay of Hinucagan, Municipality of Gandara, and island of Samar in The Philippines.

Population genetics has the potential to contribute greatly to our understanding of the epidemiology of multi-host zoonotic parasites, such as S. japonicum. Heterogeneity of schistosomes in nature is difficult to study due to the location of the adult worms within the definitive host species' blood system and the small size of the larval stages associated with transfer between hosts. We have been able to circumvent these difficulties by using single-locus molecular probes (microsatellite markers) which are present in both larval and adult worm stages of the parasites, and, for the first time, use individual larval stages which are directly collectable from natural populations for molecular epidemiological studies of the parasite, thereby also circumventing the ethical, technical and epidemiological disadvantages of laboratory passage. The data presented here, albeit as yet only on a small scale, further demonstrates the potential for host-imposed selection by laboratory mammals during isolate collection (LoVerde et al. 1985; Theron and Pointer, 1995) and highlights the need to be careful of such biases introduced by laboratory passage or collection techniques particularly in zoonotic pathogens.
Comparison of sets of cercariae from natural infections with the resultant adult worms following a single laboratory passage here reflected a general loss of alleles, most likely due to founder effects and other bottlenecking processes. This is perhaps not surprising given the recovery rate of adult worms (number of adult worms recovered in comparison to the number of cercariae used for infection which directly reflects the number of parasites that establish within the definitive host species) varied from 2·6% to 26·7% with an average of only 12%. Indeed in this study 4 isolates were lost (cercariae introduced to definitive host species but no successful establishment of parasite occurred leading to no recovery of worms and miracidia) even though they were exposed to the same laboratory-bred definitive host species, which, by highlighting the practical complexities and inconsistencies inherent in laboratory passage, suggests a further advantage of the larval approach in that all parasite isolates are available for genetic analyses without relying on successful establishment in the laboratory host.
Our data also suggest that miracidia are capable of representing all of the alleles present in the adult worms. In addition, they identify alleles generated during sexual reproduction which might in turn be characteristic of the selection pressures encountered within the definitive host species and/or related morbidity arising within the hosts due to these selected parasite genotypes. Therefore, since more alleles will be sampled for the same number of parasite samples from an individual, the analysis used here will be potentially more reliable in studying genetic substructuring within strains. However, inadequate sampling will lead to a possible loss of some alleles present in the adult population (which might not be representative of loss during sexual reproduction), hence the quantity of miracidial samples per individual species and/or geographical isolates should be kept high to accommodate as many alleles as possible. Indeed, our data from goats suggested the miracidia may cluster away from adult worms, whilst the superior rat data, in terms of where all miracidia were sampled, indicated that miracidial sampling is representative of the adult populations (as long as a sufficient miracidial sample size is analysed in order to prevent a bias). The work here will be extended to develop a recommendation for adequate sample size of miracidia that should be sampled per organism.
One of the limitations of the current analysis, nevertheless, was that all the heterogeneity and genetic differentiation tests could be performed only at an individual locus level. To obtain a realistic representation of parasite diversity it is important to perform multi-locus analysis (Rosenberg et al. 2002). However, all the 6 loci used here have been previously examined for linkage disequilibrium, and normal segregation of all the loci may be assumed since no significant linkage disequilibrium was observed between loci within individuals (Shrivastava et al. 2005). Hence, the probability of any clustering observed at the level of a single-locus being represented even at the multi-locus-genotype level is very high. Such an analysis should not overestimate any genetic structure between parasite populations, although underestimation of structures between parasite populations remains possible. Furthermore, because of the extremely low amount of DNA in each individual larva and the very high levels of cross-amplifications obtained during multiplexing these loci, locus-by locus analysis is currently the only option available for this kind of study. Current research in our laboratory is aimed at overcoming these limitations by enabling multiplexing (to allow multi-locus genotyping) of S. japonicum larval stages. Nevertheless, even considering those limitations, our results demonstrate that the technique presented here can be very successfully used for molecular epidemiological assays.
In summary, we have developed novel assays for the storage and microsatellite analysis of larval stages of human schistosomes, which negates the need for laboratory passage. This thereby circumvents the problems associated with host-induced selection and bottlenecking inherent in transfer to laboratory passage and promises a number of additional benefits. For instance, such methodology is in keeping with the ‘3Rs’ (the reduction, refinement and replacement of animal use), the cornerstone of animal experimentation ethics (Wolfensohn and Lloyd, 1998). Our methodology is also logistically easier, more reliable and less time-consuming than previous methods, allowing collection and analysis of samples within a matter of days, rather than the several months required for animal passage. Further, by using the larval stages all natural vertebrate infections will be sampled, even those that are a result of trickle exposures over time. Moreover, our assays uniquely allow us to directly address questions related to epidemiological parameters of the larval stages themselves, such as natural bottlenecking and competitive processes. For example, competition between schistosome genotypes within snails is well documented within laboratory populations and is thought to be potentially important in the evolution of schistosome virulence (Davies, Fairbrother and Webster, 2002). Likewise, temporal and spatial comparisons of natural miracidial and cercarial populations would aid in interpreting redundancy in the schistosome life-cycle, the transmission potential of host-adapted strains and the potential for intra-snail competition in the field. We therefore conclude that laboratory maintenance is often unnecessary and unadvisable for population genetic studies of multi-host parasites.
We thank Nicole Bilek and Lynne Richardson for assistance with Genescan reactions. STEP staff members from the Research Institute of Tropical Medicine, Manila, the Anhui Institute of Parasitic Diseases and the station of schistosomiasis control of Tongling, Hexian, Dangtu, Nanling and Guichi County, and Maria Van Johanson from the Danish Bilharzia Laboratory are all also thanked for their help with parasite collection. We also wish to thank staff at the Natural History Museum-London, UK, for allowing and assisting us with the temporary use of their facilities. This work was funded by a Felix scholarship (J.S.) and grants from the Royal Society (J.P.W., GR070244AiA), the Wellcome Trust (B.Z.Q. and J.P.W.) (C.M.G. and J.P.W., GR063774MA), DBL/TDR (T.P.W. and J.P.W.) and the NIH/NSF (EBjr, J.P.W., TW01582).

Table 1. Optimization of collection and storage protocol for larval stages.

Table 2. Comparison of adult worms obtained by perfusing rabbits and mice exposed to the cercariae of the same set of infected Oncomelania snails collected in Anhui province, China

Fig. 1. UPGMA phenogram depicting co-ancestry identity between Schistosoma japonicum adult worms passaged from mice and rabbits. Each population represents 20 worms from an individual definitive host.

Table 3. Comparison between cercariae collected from Cabarsan Guti, Palo, Leyte, Philippines and adult/miracidia stages of parasites obtained from laboratory mice infected with the cercariae from same snails

Table 4. Hardy-Weinberg equilibrium test (Exact test using 100000 Markov chain steps with 1000 dememorization steps in Arlequin). Expected (He) and observed (Ho) heterozygosity for each polymorphic microsatellite locus

Fig. 2. UPGMA phenogram depicting co-ancestry identity between cercariae and adult worms plus miracidial stages of Schistosoma japonicum obtained from passaging through laboratory mice. The corresponding snails had been collected from Cabarasan Guti, Palo-Leyte.

Fig. 3. UPGMA phenogram depicting co-ancestry identity between adult and miracidial stages of Schistosoma japonicum obtained from domesticated goats collected form Anhui Province of China.

Table 5. Comparison between adult and miracidial stages of parasites obtained from domesticated goats collected from Anhui Province of China.

Fig. 4. UPGMA phenogram depicting co-ancestry identity between adult and miracidial stages of Schistosoma japonicum obtained from wild rats collected from Barangay of Hinucagan, Municipality of Gandara, and island of Samar in The Philippines.

Table 6. Comparison between adult and miracidial stages of parasites obtained from wild rats collected from Barangay of Hinucagan, Municipality of Gandara, and island of Samar, Philippines