Introduction
The lichen genus Caloplaca Th. Fr. is a highly conspicuous and species-rich component of the temperate Australasian lichen flora. It is responsible for the vivid, eye-catching red, orange and yellow coloration of coastal rocks, man-made substrata such as tile roofs, stone walls and concrete, as well as tree bark and rock outcrops in natural habitats. Recent decades have seen considerable advances in the study of Caloplaca in Australia (including Tasmania), chiefly by the Ukrainian lichenologist Sergey Kondratyuk who, with collaborators, has described more than 75 species based on Australian types (Kärnefelt & Kondratyuk Reference Kärnefelt and Kondratyuk2004; Kondratyuk et al. Reference Kondratyuk, Kärnefelt, Elix and Thell2007a, Reference Kondratyuk, Kärnefelt, Elix and Thellb, Reference Kondratyuk, Kärnefelt, Elix and Thell2009a, Reference Kondratyuk, Kärnefelt, Elix and Thellb, Reference Kondratyuk, Kärnefelt, Thell and Elix2010, Reference Kondratyuk, Elix, Kärnefelt and Thell2011, Reference Kondratyuk, Elix, Kärnefelt and Thell2013a; Lumbsch et al. Reference Lumbsch, Ahti, Altermann, Amo de Paz, Aptroot, Arup, Bárcenas Peña, Bawingan, Benatti and Betancourt2011; Kantvilas & Kondratyuk Reference Kantvilas and Kondratyuk2013), culminating in a key to the more than 120 species recorded for Australia (Kondratyuk et al. Reference Kondratyuk, Elix, Kärnefelt and Thell2012). Other taxa have been added by Hafellner (Reference Hafellner1982), Kantvilas & Søchting (Reference Kantvilas and Søchting2013) and Kantvilas (Reference Kantvilas2016), to the extent that the complement of species for the region today stands at 135 taxa, of which 45 are reported for Tasmania (McCarthy Reference McCarthy2020).
Traditionally, Caloplaca has encompassed lichens with a trebouxioid photobiont, a subfruticose, placodioid, squamulose or crustose thallus, apothecial ascomata, Teloschistes-type asci, hyaline, usually polaribilocular ascospores and, in most species, orange or yellowish, K+ purple anthraquinone pigments in the thallus and/or apothecia (e.g. Fletcher & Laundon Reference Fletcher, Laundon, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009; Kantvilas Reference Kantvilas2016). However, phylogenetic studies using DNA sequence data indicated that the genus is heterogeneous (Søchting & Lutzoni Reference Søchting and Lutzoni2003; Gaya et al. Reference Gaya, Högnabba, Holguin, Molnar, Fernández-Brime, Stenroos, Arup, Søchting, van den Boom and Lücking2012; Bungartz et al. Reference Bungartz, Søchting and Arup2020) and, as a result, a large number of smaller, more natural genera have been erected (Arup et al. Reference Arup, Søchting and Frödén2013; Kondratyuk et al. Reference Kondratyuk, Jeong, Yu, Kärnefelt, Thell, Elix, Kim, Kondratiuk and Hur2013b, Reference Kondratyuk, Jeong, Yu, Kärnefelt, Thell, Elix, Kim, Kondratiuk and Hur2014a, Reference Kondratyuk, Kärnefelt, Thell, Elix, Kim, Jeong, Yu, Kondratiuk and Hurb, Reference Kondratyuk, Kärnefelt, Thell, Elix, Kim, Kondratiuk and Hur2015a, Reference Kondratyuk, Lőkös, Kim, Kondratiuk, Jeong, Jang, Oh and Hurb, Reference Kondratyuk, Lőkös, Kim, Kondratiuk, Jeong, Jang, Oh, Wang and Hur2016, Reference Kondratyuk, Lőkös, Upreti, Nayaka, Mishra, Ravera, Jeong, Jang, Park and Hur2017, Reference Kondratyuk, Persson, Hansson, Mishra, Nayaka, Liu, Hur and Thell2018a, Reference Kondratyuk, Persson, Hansson, Lőkös, Liu, Hur, Kärnefelt and Thellb), many with representatives in Australia. This new classification has not been without controversy, and has also proved unwieldy to most taxonomists working with traditional morphological and anatomical characters. Consequently, it has not been generally taken up (e.g. see Gaya et al. Reference Gaya, Fernández-Brime, Vargas, Lachlan, Gueidan, Ramírez-Mejía and Lutzoni2015; Aptroot & Cáceres Reference Aptroot and Cáceres2016; Kantvilas Reference Kantvilas2016; McCune Reference McCune2017; McCarthy Reference McCarthy2020) and it seems inevitable that, because of its easy recognition, Caloplaca in the broad sense is likely to remain in use for the foreseeable future.
Species of Caloplaca can occur on almost every conceivable lichen substratum (wood, bark, soil, humus, bryophytes, calcareous and non-calcareous rock, man-made surfaces) with the notable exception of living leaves. Almost 40 species, spanning several of the segregate genera (e.g. Athallia, Catenarina, Flavoplaca, Gyalolechia, Pachypeltis, Variolaria), are obligately or facultatively lichenicolous (Poelt & Hinteregger Reference Poelt and Hinteregger1993; Nimis et al. Reference Nimis, Poelt and Tretiach1994; Vondrák et al. Reference Vondrák, Halıcı, Güllü and Demirel2016; Diederich et al. Reference Diederich, Lawrey and Ertz2018). In general, many of these lichenicolous Caloplaca species have a comparatively wide range of hosts and their distribution appears to be determined as much by their substratum as by the taxonomy of their host lichen.
In the course of the Tasmanian Museum and Art Gallery's inaugural Expedition of Discovery, an initiative aimed at the collection and documentation of the flora and fauna from poorly studied areas of Tasmania (Baker et al. Reference Baker, Grove, de Salas, Byrne, Cave, Bonham, Moore and Kantvilas2019), a lichenicolous Caloplaca, growing on the widespread, saxicolous species Tephromela atra (Huds.) Hafellner, was discovered. No lichenicolous species of the genus are known from Australia, nor does the species closely resemble any non-lichenicolous species known in the region. After comparison of our species against the worldwide literature on lichenicolous Teloschistaceae, and to morphologically similar species from Australia, we conclude that it is new to science and describe it below.
Materials and Methods
The study is based on the collections of the new species from Tasmania and housed in the Tasmanian Herbarium (HO). For comparison with other Caloplaca taxa, we consulted reference herbarium specimens or, where these were unavailable, published species descriptions.
Anatomy and morphology
Observations of specimens were made using low-power and high-power microscopy. Thin, hand-cut sections of the apothecia were examined in a range of mounting media, including water, 10% KOH (K), Lugol's solution (I) and lactophenol cotton blue (LCB). Following the protocol described in Kantvilas (Reference Kantvilas2016), all measurements were undertaken exclusively in sections hydrated in water and then mounted in LCB. Likewise, observations of paraphyses and oil vacuoles were undertaken in LCB.
DNA extraction, PCR amplification and DNA sequencing
DNA extraction and amplification were carried out in the Mycology Laboratory of the University of Tartu (TU). Genomic DNA was extracted from ascomata using the High Pure PCR Template Preparation Kit (Roche Applied Science®), following the protocol provided by the manufacturer. We amplified three gene loci: internal transcribed spacer (nuITS) using primer pair ITS0F and LA-W (Tedersoo et al. Reference Tedersoo, Jairus, Horton, Abarenkov, Suvi, Saar and Kõljalg2008); large subunit nuclear ribosomal RNA gene (nuLSU) with LROR and LR7 (Vilgalys & Hester Reference Vilgalys and Hester1990); and mitochondrial small subunit ribosomal RNA gene (mtSSU) with mrSSU1 and mrSSU3R (Zoller et al. Reference Zoller, Scheidegger and Sperisen1999). The PCR reaction mix (25 μl) consisted of 5 μl 5× HOT FIREPol Blend Master Mix (with 10 mM MgCl2; Solis BioDyne, Tartu, Estonia), 0.5 μl of both primers (all 20 μM) and 3–8 μl of target-DNA, with the remainder being distilled water. The temperatures and time for each cycle of the polymerase chain reaction (PCR) were as follows: denaturation was set at 95 °C for 30 s; annealing at 57 °C (nuITS) or 55 °C (nuLSU, mtSSU) for 30 s; and extension at 72 °C for 60 s. A total of 36 (nuITS) and 35 (nuLSU, mtSSU) cycles were run. The PCR products were visualized on a 1% agarose gel stained with ethidium bromide, and for the purification of PCR products, 1 μl of FastAP and 0.5 μl of Exonuclease I (Thermo Scientific, Waltham, Massachusetts, USA) were added to each tube per 20 μl of the product. Both complementary strands were sequenced by Macrogen Inc. (Amsterdam, The Netherlands). The nuITS sequences were sequenced with primer pair ITS4 and ITS5 (White et al. Reference White, Bruns, Lee, Taylor, Innis, Gelfand, Sninsky and White1990), nuLSU with CTB6 (Garbelotto et al. Reference Garbelotto, Lee, Slaughter, Popenuck, Cobb and Bruns1997) and LR7, and for the mtSSU the same primers were used as for the amplification. Sequencher v.4.10.1. (Gene Codes Corp.®, Ann Arbor, Michigan, USA) was used to check, assemble and manually adjust the resulting sequence fragments. The consensus sequences were compared with those publicly available in GenBank using the ‘blastn’ algorithm (Altschul et al. Reference Altschul, Gish, Miller, Myers and Lipman1990). The newly generated DNA sequences are deposited in the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) and UNITE (Nilsson et al. Reference Nilsson, Larsson, Taylor, Bengtsson-Palme, Jeppesen, Schigel, Kennedy, Picard, Glöckner and Tedersoo2019) databases.
Phylogenetic analyses
We compiled DNA alignments for each gene, using taxon sampling that encompassed as many of the segregate caloplacoid genera as possible (Table 1). Physcia dubia and P. stellaris (Physciaceae) or Calicium viride (Caliciaceae) were included to root the phylogenies. The DNA sequences were aligned with the on-line version of MAFFT v.7 (Katoh et al. Reference Katoh, Rozewicki and Yamada2019) using default options and corrected manually with SeaView v.4.6 (Gouy et al. Reference Gouy, Guindon and Gascuel2010). The online version of Gblocks v.0.91b (Talavera & Castresana Reference Talavera and Castresana2007) was used to eliminate poorly aligned positions and divergent regions, but allowing smaller final blocks and gap positions within the final blocks. The basic statistics for all three alignments are given in Table 2.
Table 1. Taxon sampling and GenBank accession numbers of sequences of Teloschistales used in the molecular phylogenetic analyses. Newly generated sequences are in bold. Lichenicolous taxa are marked with L.

Table 2. Basic statistics for nuITS, nuLSU and mtSSU alignments of Teloschistales species in this study: number of sequences, number of nucleotide positions in original and curated (after implementation of Gblocks (Talavera & Castresana Reference Talavera and Castresana2007)) alignments, and number of variable and informative sites in curated alignment.

Alignments were analyzed using the Markov chain Monte Carlo (MCMC) and maximum likelihood (ML) approaches. The best-fit nucleotide substitution models (TIM + I + G for nuITS and GTR + I + G for nuLSU and mtSSU) were calculated over 56 models and selected based on the lowest value AIC criterion (Akaike Reference Akaike1974) with jModelTest v.2.1.6. (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012). The Bayesian analysis was performed with MrBayes v.3.2.1. (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) using the following settings: two parallel simultaneous runs over 1 million (nuITS, nuLSU) or 2 million generations (mtSSU), starting with a random tree and employing four simultaneous chains; sampling after 1000 steps. The analyses were run until convergence of the chains was confirmed by the standard deviation of split frequencies reaching 0.01, and the average Potential Scale Reduction Factor (PSRF) value was close to 1. The first 25% of saved data was discarded as ‘burn-in’. The consensus tree and posterior probabilities (PP) were calculated from the remainder. The ML analysis was run with RAxML v.8.2.12 (Stamatakis et al. Reference Stamatakis, Hoover and Rougemont2008) and inferred assuming GTR + G as the nucleotide substitution model. Branch support was calculated by rapid bootstrapping over 1000 pseudoreplicates. All analyses were implemented on the CIPRES Science Gateway v.3.3 (Miller et al. Reference Miller, Pfeiffer and Schwartz2010). Those clades with posterior probabilities (PP) ≥ 0.95 and bootstrap values (BS) ≥ 0.75 were regarded as significantly supported. We present only the nuITS tree (Fig. 1) because here the number of taxa was largest (see Table 2). The phylogenetic tree was visualized and edited using FigTree v.1.4.2 (Rambaut Reference Rambaut2014). Adobe Illustrator CS3® was used for artwork.

Fig. 1. The rDNA ITS-based consensus tree derived by the Bayesian method, showing the position of Caloplaca tephromelae within the Teloschistales. The branches with Bayesian posterior probabilities (PP) ≥ 0.95 and maximum likelihood bootstrap values (BS) ≥ 75 indicated above the branches are considered as supported and marked with a thicker line. The supported clades, and clades corresponding to generic rank according to various authors, are collapsed; numbers in brackets after taxon names indicate number of sequences in this clade. Lichenicolous taxa are indicated with ‘L’ and apparently incorrectly identified sequences with ‘*’.
Results
No sequence identical to that of the new species was found from the nucleotide databases, but the closest match was always a member of the Teloschistaceae. The percentage identity with the closest taxon ranged from 88% (nuITS) to 95% (mtSSU) and 97% (nuLSU). The phylogenetic analyses did not resolve the position of the new Caloplaca species because whereas the nuITS analysis showed a sister relationship with the Yoshimuria clade (PP = 1, but BS = 64), the mtSSU analysis suggested a relationship with Marchantiana occidentalis (PP = 1, BS = 95). Supported relationships were not found in the nuLSU analysis. The phylogenetic analyses did not reveal any relationship with Caloplaca (Erichansenia) epithallina, a taxon which inhabits Tephromela thalli, nor with any other lichenicolous species of Caloplaca for which sequences were available (Fig. 1, Table 1). Although we prefer to retain the generic name Caloplaca in the broad sense, we acknowledge that groups of related taxa have been accorded generic rank by various authors. Accordingly, we present these in Fig. 1 as a means of illustrating where the various species groups are positioned in our phylogeny. The results confirm that the lichenicolous habit in Caloplaca s. lat. has arisen several times in the phylogeny (indicated with an ‘L’ after the relevant taxa) and is not confined to any particular group of related taxa. Sequences AF279885, annotated as Caloplaca cerina, and KC179123 as Filsoniana kiamae, are apparently incorrectly identified (Fig. 1).
Taxonomy
Caloplaca tephromelae Kantvilas, Suija & Motiej. sp. nov.
MycoBank No.: MB 838715
Species lichenicola, thallum Tephromelae incolens, apotheciis lecanorinis vel zeorinis, margo albido-griseo, pigmentum aurantiacum destituto, hymenio non-insperso, paraphysibus oleo-vacuolas deficientibus, ascosporis 10–14 μm longis, 5–8 μm latis, septo 5–8 μm crasso recognita.
Typus: Australia, Tasmania, Wind Song Property, northern rim of Callitris Gully, 42°20′55″S, 147°55′03″E, 60 m alt., on thallus of Tephromela atra, growing on dolerite outcrops in degraded rough pasture, 22 February 2019, G. Kantvilas 26/19 (HO—holotypus; BILAS, TUF091318—isotypi). GenBank Accession nos: MW483077 (nuLSU), MW483076 (mtSSU). DNA barcode/reference (nuITS) sequence from isotype: GenBank MW485494 / UNITE UDB0778961. UNITE SH3597440.08FU.

Fig. 2. Caloplaca tephromelae habit, showing the small, lecanorine apothecia with an orange-brown disc, growing on the whitish thallus of Tephromela atra (with large lecanorine apothecia with a black disc). Scale = 2 mm.

Fig. 3. Caloplaca tephromelae anatomy. Paraphyses, Teloschistes-type asci with amyloid parts stippled and ascospores (semi-schematic). Scale = 10 μm.
Thallus whitish, areolate, occurring as small islands, bordered by a dark band of prothallus, within the thallus of Tephromela atra and T. granularis, or ±autonomous in close association with these species; medulla patchily I+ blue.
Apothecia 0.2–0.7 mm wide, lecanorine to zeorine, scattered over the thallus of the host or crowded together in discrete clusters, roundish or a little distorted due to mutual pressure, sessile, basally constricted; disc orange to dull orange-brown, matt, somewhat coarsely pruinose, persistently plane or a little undulate; thalline margin dull whitish grey, sometimes a little bluish grey to brownish, usually entirely enveloping the apothecia but at times crenulate, incomplete and mainly around the apothecium base, in section 50−80 μm thick, inspersed with crystals that fluoresce white in polarized light and dissolve in K; photobiont trebouxioid, cells ±globose, 6−12 × 5−10 μm, extending ±continuously beneath the hymenium but absent from the outermost 15−20 μm of the margin; proper excipulum either obscured by the thalline margin or seen as a thin, rather glossy dark brown rim between the thalline margin and the disc, in section 20−50 μm thick, cupulate, poorly differentiated from the subhymenium, composed of intertwined, short-celled hyphae with cells 3−6 μm wide. Subhymenium hyaline, 40–100 μm thick in the central part, usually inspersed with minute oil droplets. Hymenium 70–90 μm thick, hyaline, not inspersed, overlain by a dense band of golden yellow crystals 10−15 μm thick that fluoresces orange-yellow in polarized light; paraphyses 1.5–2 μm thick, lacking oil vacuoles, sparsely branched, with apices mostly moniliform and expanding to 2.5–4 μm at the apices; asci 8-spored, 45–60 × 13–20 μm. Ascospores polaribilocular, ellipsoid, 10–14 × 5–8 μm; septum 5–8 μm.
Pycnidia not found.
Chemistry
Thallus and apothecial margin K−; apothecial disc K+ crimson (anthraquinone pigments); composition of secondary compounds not analyzed.
Etymology
The specific epithet refers to the host of the new lichen.
Distribution and ecology
The major host species, Tephromela atra is very widespread and common in Tasmania, and ranges from littoral to alpine altitudes. It occurs on a wide range of rock types, usually in exposed situations, in forest, heathland, grassland and highly modified agricultural environments. The sorediate T. granularis Kantvilas, on which the new species has also been observed, is more restricted and occurs on dolerite outcrops in low rainfall areas, mostly in open eucalypt forest or in degraded, heavily grazed scrubby pasture. Both host taxa are well represented in herbaria; all available collections (>100) were examined but failed to reveal any further material of the new species. Thus Caloplaca tephromelae is still known only from the type locality where it grew on large dolerite outcrops in a highly degraded, roughly-cleared sheep pasture. The first collection made was rather fortuitous, but on revisiting the site the new species was found to be abundant, although extremely localized on just a small number of outcrops. The boulders on which the new species occurs support a diverse suite of foliose and crustose lichens. Major species present include Caloplaca (Nevilleiella) lateritia (Taylor) Zahlbr., Carbonea latypizodes (Müll. Arg.) Knoph & Rambold, Flavoparmelia haysomii (C. W. Dodge) Hale, Lecanora farinacea Fée, Lecidea atromorio C. Knight, Monerolechia badia (Fr.) Kalb, Paraporpidia leptocarpa (C. Bab. & Mitt.) Rambold & Hertel, Punctelia subrudecta (Nyl.) Krog, Ramboldia petraeoides (Nyl. ex C. Bab. & Mitt.) Kantvilas & Elix, Rhizocarpon geographicum (L.) DC., R. reductum Th. Fr. and numerous species of Xanthoparmelia.
Additional specimen examined
Australia: Tasmania: type locality, 2017, G. Kantvilas 309/17 (HO) [on T. atra and T. granularis].
Discussion
Of the approximately 40 lichenicolous Caloplaca (in the wide sense) species, sequences are available only for about one third of them (Table 1) and none of these revealed any close relationship with C. tephromelae (Fig. 1). Lichenicolous Caloplaca species display a wide range of characters with respect to the morphology and coloration of the thallus and apothecia, ascospore form and size, as well as other features. Consequently, we narrowed our detailed comparison of salient morphological and anatomical features to those lichenicolous species with lecanorine apothecia and lacking K+ crimson, anthraquinone pigments in the thallus (Table 3).
Table 3. Comparison of lichenicolous species of Caloplaca with a grey or inapparent thallus and lecanorine apothecia. ‘Nd’ indicates that no data were given in the reference.
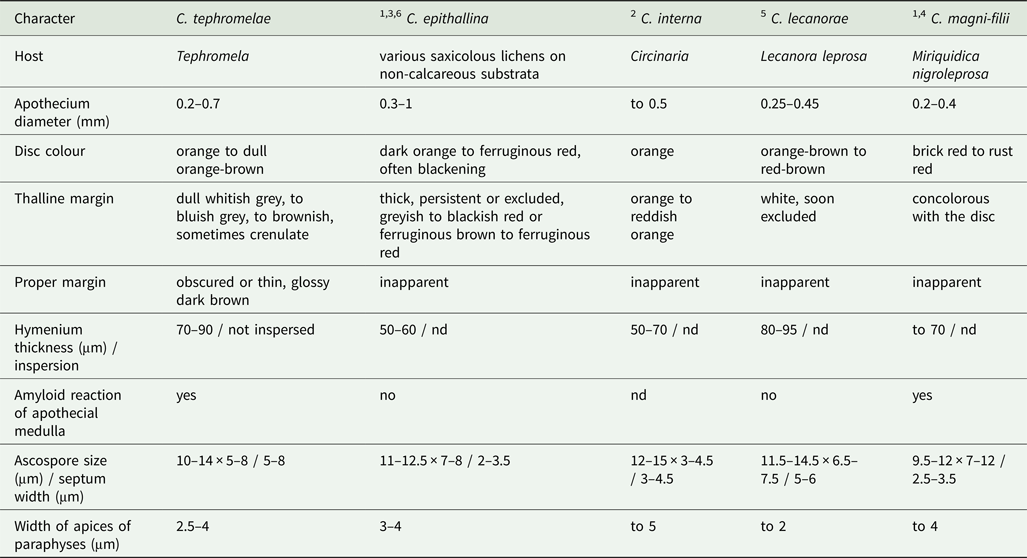
1Hansen et al. (Reference Hansen, Poelt and Søchting1987); 2Nimis & Poelt (Reference Nimis and Poelt1987); 3Øvstedal et al. (Reference Øvstedal, Tønsberg and Elvebakk2009); 4Poelt (Reference Poelt1958); 5Seavey & Seavey (Reference Seavey and Seavey2012); 6Søchting et al. (Reference Søchting, Balschmidt Lorentsen and Arup2008)
Of these morphologically similar, lichenicolous species, Caloplaca epithallina Lynge is the only one for which sequences were available, and these indicated that it is not closely related to C. tephromelae (Fig. 1). It also differs from C. tephromelae in that its apothecial margin is concolorous with the disc, the hymenium is thinner (to 60 μm thick) and the ascospores, although of a similar size to those of C. tephromelae, have a septum only 2−3.5 μm thick (Hansen et al. Reference Hansen, Poelt and Søchting1987; Øvstedal et al. Reference Øvstedal, Tønsberg and Elvebakk2009). This Northern Hemisphere species is known to occur on Tephromela, as well as on some other saxicolous crustose and foliose lichens.
The most similar species to C. tephromelae morphologically is C. lecanorae F. Seavey & J. Seavey, described from Lecanora leprosa Fée in Florida. This species differs in having smaller apothecia (0.25−0.45 mm wide), with a thalline margin that is soon excluded and an inapparent proper margin (Seavey & Seavey Reference Seavey and Seavey2012), as well as in the lack of a medullary iodine reaction (F. Seavey, personal communication; Table 3). Despite our efforts, material of this species for molecular analysis and comparison could not be obtained. Two further lichenicolous Caloplaca species with lecanorine apothecia lack anthraquinone pigments in the thallus: Caloplaca interna Poelt & Nimis differs in having a narrower ascospore septum (2–3.5 μm), wider paraphyses tips (to 5 μm) and a thalline apothecial margin containing anthraquinone pigments, and occurs on Circinaria species on calcareous substrata (Nimis & Poelt Reference Nimis and Poelt1987); Caloplaca magni-filii differs by its strongly convex apothecia with darker apothecial discs, the thalline margin containing anthraquinone pigments, and the broadly ellipsoid to rounded ascospores with a narrow septum (2.5–3.5 μm), and occurs on Miriquidica nigroleprosa (Poelt Reference Poelt1958; Hansen et al. Reference Hansen, Poelt and Søchting1987). Both species occur only in the Northern Hemisphere.
Although the genus Caloplaca is very species-rich in Australia and Tasmania, no lichenicolous species have been reported so far from this region. Amongst the autonomous taxa, the critical characters of the new taxon, notably the whitish thallus lacking anthraquinones and the lecanorine to zeorine apothecia, are also uncommon and seen only in the corticolous C. bastowii S. Y. Kondr. & Kärnefelt and the saxicolous C. kilcundaensis S. Y. Kondr. & Kärnefelt. Both of these taxa differ from C. tephromelae by having significantly larger apothecia (to 1.2 mm wide), a hymenium and subhymenium densely inspersed with oil droplets, and ascospores with a narrower septum (at most to 4 μm wide) (Kantvilas Reference Kantvilas2016). The latter differs further in having a thallus with a distinct brownish tinge and apothecia that are mostly biatorine and only secondarily develop a thalline margin.
Acknowledgements
We thank Jean Jarman for the photograph and preparing the figures for publication. Frederick Seavey is thanked for supplying additional details for Caloplaca lecanorae. Financial support for AS was provided by the European Regional Development Fund (Centre of Excellence EcolChange) and an Estonian Research Council grant (PRG1170). Rasmus Puusepp (Tartu) is thanked for laboratory work. Fieldwork in Tasmania where the new species was discovered was supported by Jane and Tom Teniswood (Wind Song, Tasmania) and the Friends of the Tasmanian Museum and Art Gallery.
Author ORCIDs
Gintaras Kantvilas, 0000-0002-3788-4562; Jurga Motiejūnaitė, 0000-0002-6949-1990; Ave Suija, 0000-0003-3784-9414.









