INTRODUCTION
The superfamily Brachylaimoidea Allison, 1943 represents a group of digeneans with poorly understood phylogenetic relationships. Brachylaimoidea consists of parasites of birds and mammals, rarely of amphibians and reptiles, affecting domestic animals (Balozet, Reference Balozet1937), poultry (Harkema, Reference Harkema1939; Alicata, Reference Alicata1940; Hodasi, Reference Hodasi1967; Baruš et al. Reference Baruš, Ryšavý and Groschaft1969; de Faria Duarte, Reference de Faria Duarte1980), wild game birds (Joyeux et al. Reference Joyeux, Baer and Timon-David1934; Gvozdev, Reference Gvozdev1953) and occasionally humans (Butcher et al. Reference Butcher, Talbot, Norton, Kirk, Cribb, Forsyth, Knight and Cameron1996, Reference Butcher, Parasuramar, Thompson and Grove1998; Butcher and Grove, Reference Butcher and Grove2001). Brachylaimoidea have a very short or absent oesophagus, gonads posterior to the ventral sucker, short excretory vesicle, two long lateral excretory canals and numerous flame cells. Sporocysts are branched, and cercariae lack the tail or their tail is only poorly developed (Gibson et al. Reference Gibson, Jones and Bray2002). Brachylaimoidea, along with Dicrocoeliidae, are the only trematodes that are capable to complete their life cycle outside of wetlands, sometimes even in xerophilic, arid habitats; only the life cycle of Leucochloridiomorphidae is completed in an aquatic environment (Sirgel et al. Reference Sirgel, Artigas, Bargues and Mas-Coma2012).
Some genera of Brachylaimoidea, such as Leucochloridium and Brachylaima, are broadly distributed and relatively well known. However, the adults of Brachylaima spp. and Leucochloridium spp. are difficult to identify to species because of few autapomorphic characters and due to the presence of abundant eggs in gravid worms, which frequently obscure internal organs (Casey et al. Reference Casey, Bakke, Harris and Cable2003). Taxonomic position of several other genera within this family is even more complicated as the genera Michajlovia, Urorygma and Zeylanurotrema are still considered incertae sedis. Olson et al. (Reference Olson, Cribb, Tkach, Bray and Littlewood2003) suggested that the Brachylaimidae is a paraphyletic group with the Leucochloridiidae nested. Or if Zeylanuratrema is its own family within the superfamily, then Brachylaimidae and Leucochloridiidae are sister taxa. Despite several previous molecular phylogenetic and systematic studies ranging from family level systematics (Olson et al. Reference Olson, Cribb, Tkach, Bray and Littlewood2003; Sirgel et al. Reference Sirgel, Artigas, Bargues and Mas-Coma2012) to the differentiation among closely related species (Bakke, Reference Bakke1978; Machalska, Reference Machalska1978; Casey et al. Reference Casey, Bakke, Harris and Cable2003; Olson et al. Reference Olson, Cribb, Tkach, Bray and Littlewood2003; Iwaki et al. Reference Iwaki, Okamoto and Nakamori2009; Locke et al. Reference Locke, Lapierre, Byers, Proctor, McLaughlin and Marcogliese2012; Zhukova et al. Reference Zhukova, Prokhorova, Tsymbalenko, Tokmakova and Ataev2012; Rząd et al. Reference Rząd, Hofsoe, Panicz and Nowakowski2014), phylogenetic relationships within the Brachylaimoidea are still prevalently based on morphology and life cycles, with conflicting opinions on the number of species and their synonymization.
In the present work, we address the evolutionary relationships of central European species of the Brachylaimoidea based on their molecular and comparative morphological analysis. We perform first conclusive phylogenetic analysis of the taxonomic position of central European Brachylaimoidea based on four independent nuclear and mitochondrial DNA loci. We also provide comparative measurements of the examined central European Brachylaimoidea, and address their tissue specificity and host-specific prevalence based on the extensive cohort of birds examined in years 1962–2015.
MATERIAL AND METHODS
Sampling
For the purpose of prevalence assessment, we examined over 17 000 individuals of 240 bird species for the presence of helminths of the Brachylaimoidea in years 1962–2015 (det. & coll. J. Sitko). All examined birds were collected in the Czech Republic (48°39′N–50°59′N, 12°19′E–18°29′E), primarily in the eastern parts of the country as specified in detail in Table S1.
For the purpose of phylogenetic analyses, we examined representative specimens of the Brachylaimoidea collected in the Czech Republic (Záhlinice 49·17°N, 17·28°E, and those delivered to the rescue station Bartošovice 49·67°N, 18·05°E from its vicinity) and Poland (Dabie lake near Szczecin 53·42°N, 14·61°E, Kopan 54·46°N, 16·43°E and Choczewo 54·73°N, 17·90°E) from Sep-2010 to Apr-2015. All helminths were obtained from birds that were submitted for deposition in the Comenius Museum in Přerov, Czech Republic (Czech specimens) or in the University of Szczecin (Polish specimens). All birds were already dead from various causes when they were received by the institutions. The list of individuals examined is provided in Table 1.
Table 1. New sequences of members of the Brachylaimoidea, collected from the Czech Republic and Poland, generated throughout the course of this study.
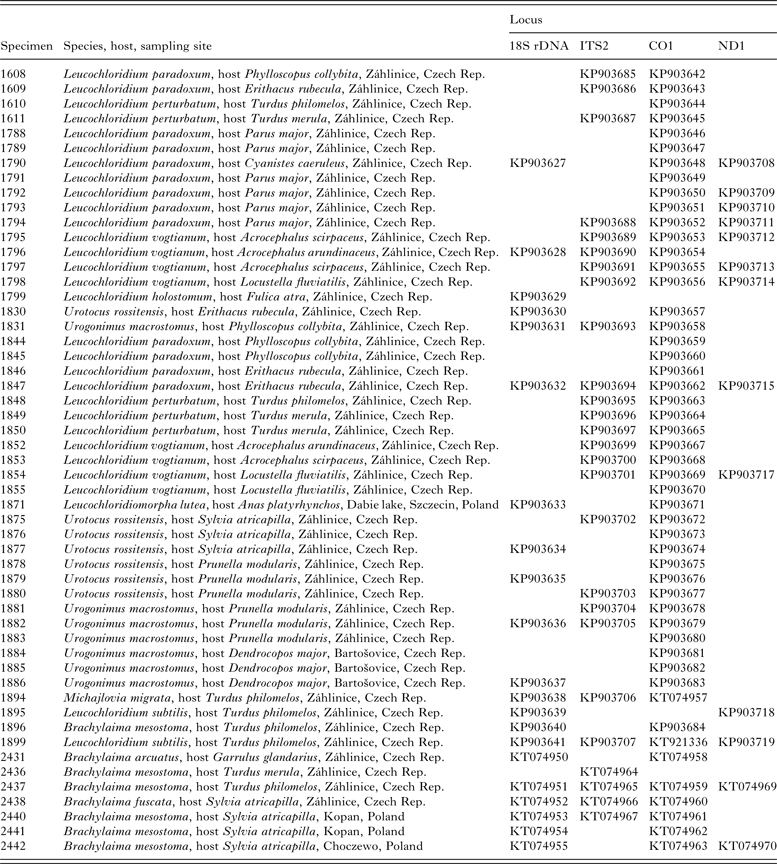
NCBI GenBank accession numbers are indicated.
For the morphological analyses, we stained representative specimens in Semichon's carmine, dehydrated by alcohol series, and mounted in Canada balsam. For the phylogenetic analyses, we fixed and stored specimens in 96% ethanol. Dimensions are shown in μm as range (mean ± s.d.). All other data are shown as mean ± s.d. unless stated otherwise.
DNA extraction, amplification and sequencing
We extracted and amplified DNA as described (Heneberg et al. Reference Heneberg, Rojas, Bizos, Kocková, Malá and Rojas2014), using the primers targeting nuclear 18S rDNA and ITS2 loci, and mitochondrial CO1 and ND1 loci (Table 2). The new primer (JB4·5-in) was designed using Primer3 (http://frodo.wi.mit.edu/primer3, accessed on 12-May-2014) based on conserved CO1 sequences of Leucochloridium spp. obtained throughout the course of this study. The DNA amplicons were purified using USB Exo-SAP-IT (Affymetrix, Santa Clara, CA) and were subjected to bidirectional Sanger sequencing using ABI 3130 DNA Analyser (Applied Biosystems, Foster City, CA). The resulting consensus DNA sequences were submitted to the GenBank database under the accession numbers KP903627-KP903719, KT074950-KT074970 and KT921336 (Table 1).
Table 2. Primers used for the amplification and sequencing of mitochondrial and nuclear DNA loci in the Brachylaimoidea.

Alignments and phylogenetic analyses
Newly generated sequences, sequences obtained from NCBI GenBank as of 8-Mar-2015, and sequences of corresponding outgroups were aligned by ClustalW (gap opening penalty 7, gap extension penalty 2 for both pairwise and multiple alignments, DNA weight matrix IUB, transition weight 0·1) as implemented in the program MEGA5. We manually corrected the alignments for any inconsistencies. We trimmed the aligned sequences, and removed short-length sequences from the alignments; only trimmed sequences were utilized for further analyses. The trimmed 18S rDNA locus (partial SSU rRNA coding sequence) corresponded to nt. 27–714 (688 bp) of Leucochloridium perturbatum AY222087 (Table S2). The trimmed ITS2 locus (partial ITS2 sequence) corresponded to nt. 606–945 (340 bp) of L. perturbatum JF331664 (Table S3). The trimmed CO1 locus (partial CO1 coding sequence) corresponded to nt. 160–329 (170 bp) of Renylaima capensis HE663454 (Table S4). The trimmed ND1 locus (partial ND1 coding sequence) corresponded to nt. 1–400 (400 bp) of Diplostomum phoxini AY386168 (Table S5).
Maximum likelihood fits of 24 nucleotide substitution models were performed as described (Řezáč et al. Reference Řezáč, Gasparo, Král and Heneberg2014), with all sites used for the analyses, including gaps. For each model, we calculated the Bayesian information criterion, Akaike information criterion (corrected) and maximum likelihood values. For the 18S rDNA locus, we analysed 27 sequences with a total of 691 positions in the final dataset (Table S6). For the ITS2 locus, we analysed 33 sequences with a total of 440 positions in the final dataset (Table S7). For the CO1 locus, we analysed 52 sequences with a total of 170 positions in the final dataset (Table S8). For the ND1 locus, we analysed 14 sequences with a total of 400 positions in the final dataset (Table S9). We used the best fit models (Tables S6–S9) for the phylogenetic analyses. We employed the bootstrap procedure at 1000 replicates. For the tree inference, we used nearest-neighbour-interchange as the maximum-likelihood heuristic method of choice, and the initial tree was formed by the neighbour joining algorithm.
We used the maximum-likelihood method to estimate inter- and intrasite evolutionary divergence in central European species of the Brachylaimoidea. We calculated the number of base differences per site by averaging over all sequence pairs between groups (Distance) ± s.e., and employed the bootstrap procedure at 1000 replicates. The model, used to estimate inter- and intrasite evolutionary divergence based on the 18S rDNA locus was identical with that used to construct the respective tree. However, to analyse the ITS2 locus, we employed the model identical with that used to construct the respective tree but without correction for the evolutionarily invariable sites, and to analyse the CO1 and ND1 loci, we employed the Tamura–Nei model (Tamura and Nei, Reference Tamura and Nei1993) with the non-uniformity of evolutionary rates among sites modeled by using a discrete Gamma distribution (+G) with 5 rate categories, because the Hasegawa–Kishino–Yano model does not allow the calculation of inter- and intrasite evolutionary divergence by the program used (Tamura et al. Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).
To corroborate the data obtained with maximum likelihood analysis, we employed Bayesian inference. To infer the tree topologies using a Bayesian approach, we converted the ClustalW alignments generated in MEGA5 to the Nexus format in Mesquite 3·04, manually adjusted the accessory information, and performed the Bayesian analysis in MrBayes 3·2·5, using the mixed model of nucleotide substitution. Bayesian analysis included four Monte Carlo Markov chains for 2 000 000 generations, and trees sampled every 1000th generation for CO1 and 18S, and less for the other two loci analysed as long as the average standard deviation of split frequencies did not exceed 0·01. We discarded the first 25% of samples as burn-in. After discarding the burn-in samples, we used the remaining data to generate a 50% majority-consensus tree with the posterior probabilitis of branches indicated. We visualized the resulting trees in FigTree 1·4·2. We obtained the following summary statistics for the for analyses performed: average standard deviation of split frequencies 0·0057–0·0077, maximum standard deviation of split frequencies 0·018–0·025, average potential scale reduction factor 1·000–1·001, and maximum potential scale reduction factor 1·003–1·017.
RESULTS
Central European species of the superfamily Brachylaimoidea
The examined birds were infected by five Leucochloridium spp. (L. holostomum, L. paradoxum, L. perturbatum, L. subtilis and L. vogtianum), Urotocus rossitensis, Urogonimus macrostomus, Michajlovia migrata, Leucochloridiomorpha lutea and three Brachylaima spp. (B. arcuatus, B. fuscata and B. mesostoma). We also found Amblosoma exile, but its available specimens were not suitable for successful DNA isolation and amplification.
Phylogenetic analyses
Leucochloridiidae
Maximum-likelihood analysis revealed the existence of distinct, well-defined clusters corresponding to particular morphologically poorly differentiated species, but only partly following currently accepted higher taxonomic units (Figs 1 and 2). The highest resolution was provided by the ND1 locus (Fig. 2B); however, the primers used to amplify this locus did not work for all species. The CO1 and ITS2 loci also provided good resolution at the species level, whereas the less variable 18S rDNA locus was helpful to elucidate the assignment to higher taxonomic units and sequences of this locus were identical in some closely related species.
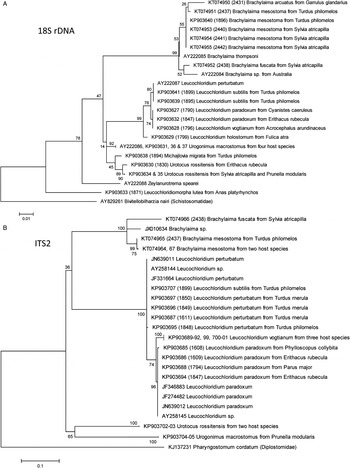
Fig. 1. Maximum-likelihood analysis of sequences of nuclear DNA loci [18S rDNA (A) and ITS2 (B)] of Brachylaimoidea.

Fig. 2. Maximum-likelihood analysis of sequences of mitochondrial DNA loci [CO1 (A) and ND1 (B)] of Brachylaimoidea.
Maximum-likelihood analysis confirmed the existence of five well-defined Leucochloridium spp. In addition to the L. paradoxum and L. perturbatum analysed previously (Rząd et al. Reference Rząd, Hofsoe, Panicz and Nowakowski2014), our data have corroborated L. holostomum, L. subtilis and L. vogtianum as separate species. The genus Leucochloridium represented a monophyletic group divided into three clades, represented by: (1) L. vogtianum and L. paradoxum, (2) L. perturbatum and L. subtilis and (3) by a more basal L. holostomum. The evolutionary divergence of all the four loci tested (Table 3) strongly supported the validity of the four species tested. Only the examined 18S rDNA locus was not sufficient to distinguish between species of the above clades. But the CO1, ITS2 and particularly the ND1 loci were all sufficient to differentiate between L. vogtianum and L. paradoxum, and ND1 was sufficient to differentiate between L. perturbatum and L. subtilis (Figs 1 and 2).
Table 3. Estimates of the intra- and interspecific evolutionary divergence of the Brachylaimoidea from the Czech Republic and Poland.

The estimates are based on sequences of the 18S rDNA, ITS2, CO1 and ND1 DNA loci. Distance: The number of base differences per site generated by averaging over all sequence pairs between groups.
Morphologic data suggest that the subfamily Leucochloridiinae is represented by the genera Urotocus, Urogonimus and Leucochloridium. However, there is no DNA-based support to retain Urotocus in Leucochloridiinae as revealed by the analysis of both the conserved (18S rDNA) and hypervariable (ITS2, CO1) DNA loci (Figs 1 and 2).
The genus Urogonimus was represented in our analyses by a single species with noticeable host species-specific intraspecific evolutionary divergence (Table 3) suggesting that host species specialization occurs in this species. There is no DNA-based support to retain Urogonimus in Leucochloridiinae as revealed by the analysis of both the conserved (18S rDNA) and hypervariable (ITS2, CO1) DNA loci (Figs 1 and 2).
Brachylaimidae
At a time of analysis, the NCBI database contained two CO1 sequences of R. capensis (Sirgel et al. Reference Sirgel, Artigas, Bargues and Mas-Coma2012). Because R. capensis was clustering with Brachylaima spp., we propose to re-classify R. capensis Sirgel and Mas-Coma (Reference Sirgel and Mas-Coma2010) as Brachylaima capensis (Sirgel and Mas-Coma, Reference Sirgel and Mas-Coma2010).
The newly defined genus Brachylaima represented a monophyletic group; the hitherto sequenced specimens did not segregate into any clades. All the sequenced species were well defined, only the 18S rDNA suggested that B. arcuatus represents an ingroup of B. mesostoma (Fig. 1A), but 18S rDNA analysis provided only shallow support at the species level, its bootstrapping revealed negligible significance of the prediction of B. arcuatus position (38%), and the analysis of CO1 rejected it at all (Fig. 2A). The combined data suggest six hitherto sequenced Brachylaima spp., represented by the Central European species B. mesostoma, B. fuscata and B. arcuatus, B. thompsoni sequenced earlier (Olson et al. Reference Olson, Cribb, Tkach, Bray and Littlewood2003), B. capensis known formerly as Renylaima capensis, and an unidentified Brachylaima sp. from Australia. More sequences of hypervariable loci need to be included in further analyses to elucidate the hierarchy within the genus Brachylaima.
Leucochloridiomorphidae
We provided the first sequences of a member of this family, namely L. lutea. This species likely has poorly conserved primer annealing sites, and we managed to obtain only 18S rDNA and CO1 of a single specimen of this species. Analysis of 18S rDNA suggested Leucochloridiomorphidae as a family basal to other Brachylaimoidea (Fig. 1A). Analysis of CO1 was inconclusive, forming a separate clade together with M. migrata. The analysis of L. lutea DNA thus did not reveal any inconsistencies in the taxonomic position of Leucochloridiomorphidae.
Panopistidae
We were the first to sequence the DNA of M. migrata, considered as a genus incertae sedis within Brachylaimoidea by Gibson et al. (Reference Gibson, Jones and Bray2002), suggested to be included in Leucochloriinae in its original description by Pojmańska (Reference Pojmańska1973), and listed among Panopistidae in Fauna Europaea (Gibson, Reference Gibson2015). The DNA sequencing data are also inconclusive. Maximum-likelihood analysis of the 18S rDNA locus suggested M. migrata as a close relative of U. rossitensis and another species incertae sedis, Zeylanurotrema spearei (Fig. 1A). Analysis of the CO1 provided only low resolution, placing M. migrata close to L. lutea, but with poor bootstrap support for such conclusion (Fig. 2A). Sequences of other Panopistidae species are unavailable. Based on our data, we can reject the recognition of M. migrata as a member of Leucochloridiinae sensu stricto (as defined above in this paper) or Brachylaimidae. Further research is needed to confirm its placement in Panopistidae or its re-classification.
Previously sequenced species of Brachylaimoidea
We included in our analyses the specimens of Glaphyrostomum sp. (NCBI Acc. No. FJ713138; Brachylaiminae) and Z. spearei (AY222088; species incertae sedis). The CO1 sequence of Glaphyrostomum sp. did not cluster with any Brachylaimidae (Fig. 2A). The 18S rDNA sequence of Z. spearei formed a long branch basal to all Brachylaimoidea except L. lutea (Fig. 1A).
We corroborated the data obtained with maximum-likelihood analysis by inferring the tree topologies using a Bayesian approach. The Bayesian approach confirmed all the key conclusion of the maximum-likelihood analyses, and the still questionable classification of Leucochloridiomorpha and Michajlovia (Figs S1–S4).
Ecology of Brachylaimoidea
The Brachylaimoidea display strong species-specific differences in host-specific prevalence (Table 4) and intensity of infection (Table 5). Brachylaima arcuatus was an intestinal parasite of only Garrulus glandarius, and was present at 10% prevalence. Brachylaima fuscata was found in the intestine of gamefowl, doves, pigeons and passeriform birds, rarely also in birds of prey and owls. Its prevalence was low in all examined hosts; we found multiple host specimens positive for this species only in Sturnus vulgaris (relative prevalence 3·0%), Pica pica (1·8%) and Buteo buteo (0·4%) (Table 4). Brachylaima mesostoma was found within the intestine of passeriform birdsFootnote 1 , characteristically in turdids, in which the prevalence reached up to 10%. Michajlovia migrata is a short-living intestinal parasite of passeriform birds of the family Turdidae, and was found by us only during the first post-migration month of thrushes. The intensity of infection reached only 1–2 individuals per infected host. Leucochloridium holostomum was found only in the large intestine of gruiform birds of the family RallidaeFootnote 2 , with particularly high prevalence in Porzana porzana (three out of four examined P. porzana were positive for this species, Table 4). Leucochloridium paradoxum was found in the large intestine, cloaca and bursa of Fabricius of passerine birds and waders, being particular for canopy birds, but with the host spectrum overlapping with L. perturbatum; we found its highest prevalence in Poecile palustris (18%) and Parus major (9%). Leucochloridium perturbatum was found in the large intestine and cloaca of passerine birds, particularly thrushes, and waders, rarely in other birds when they occasionally feed on molluscs; we found its highest prevalence in Turdus merula (24%) and Turdus philomelos (19%). Leucochloridium subtilis was an extremely rare parasite in the large intestine, cloaca and bursa of Fabricius of passerine birds; we recorded only three cases of infection, of them two in thrushes and one in Phylloscopus sibilatrix. Leucochloridium vogtianum was found in the cloaca and rectum of passerine birds associated with wetlands; we found the highest prevalence in Acrocephalus scirpaceus (5%) and other reed and grass warblers (Table 4). Urogonimus macrostomus was found in the cloaca and rectum of passeriform, piciform and charadriiform birds, but without any obvious core species. In central Europe, the intermediate hosts of U. macrostomus do not occur in lowlands, thus the findings from lowland floodplain forests are limited to the migration and winter periods. Urotocus rossitensis is a very rare parasite of the bursa of Fabricius and cloaca during summer and autumn, as found also by Okulewicz (Reference Okulewicz1991). Due to the winter atrophy of bursa of Fabricius in small passerines, U. rossitensis relocates to the large intestine for the winter period. Adults have a long lifespan during which they modify the section of intestine in which they reside, and modify, in part, their body shape. Note that body shape, dimensions and location in host change during U. rossitensis lifespan, the measurements presented in this study refer to adults obtained in the spring; key differences are listed in footnotes. Adults are sensitive to pressure, and their tegument can easily crack during slide mounting. The host range includes various passeriform birds, with the highest prevalence noticed in Prunella modularis (3%). Leucochloridiomorpha lutea is a rare parasite of the bursa of Fabricius in ducks, with up to 6% prevalence in juveniles of Anas spp. and Aythya spp., but nearly absent in adult birds.
Table 4. Host- and age-specific prevalence of the Brachylaimoidea species in the Czech Republic in years 1962–2015.

a Leucochloridiomorpha lutea occurs in juveniles only, thus the relative share of positive host birds was calculated from the number of juveniles examined excluding the adults of Anas platyrhynchos and Aythya nyroca. The only exception was Anas crecca, where we noticed the only finding of L. lutea in an adult individual, when examining 40 adults and 32 juveniles of A. crecca.
Table 5. Intensity of infection by the Brachylaimoidea species in the Czech Republic in years 1962–2015.

Comparative morphology of Brachylaimoidea
As most of the species discussed in this study were not subjected to DNA analysis previously, we complement here the DNA analyses with measurements of key morphologic features of the analysed species as identified by the combined morphologic and genetical approach (Tables 6 and 7) and provide representative figures of each species analysed (Figs 3 and 4). Brachylaima arcuatus (Fig. 3A) typically has ventral sucker larger than oral sucker; body length up to 7000; tegument spined only up to level of anterior testes; bursa cirri large, up to 300; eggs large, up to 36 × 24. Brachylaima fuscata (Fig. 3B) has ventral sucker larger than oral sucker; body length up to 4000; tegument spined only in anterior half of body; bursa cirri up to 200. Brachylaima mesostoma (Fig. 3C) has suckers of equal size; body length 1500–2200; whole tegument spined, including its posterior part. All central European Brachylaima spp. have testes arranged diagonally; vitelline follicles extend from ventral sucker to anterior half of anterior testis; dorsal, lateral and ventral to caeca, few follicles invading slightly intercaecal space.

Fig. 3. Representative microphotographs of Brachylaima arcuatus (A), B. fuscata (B), B. mesostoma (C), Michajlovia migrata (D), Urogonimus macrostomus (E) and Urotocus rossitensis (F) stained in Semichon's carmine. (A) B. arcuatus, host: Garrulus glandarius, adult male, sampling site and date: Klec, Czech Republic, 9-Mar-1968. (B) B. fuscata, host: Sturnus vulgaris, adult female, sampling site and date: Přerov, Czech Republic, 18-Oct-2000. (C) B. mesostoma, host: Sylvia atricapilla, adult female, sampling site and date: Záhlinice, Czech Republic, 30-Aug-1987. (D) M. migrata, host: Turdus merula, adult male, sampling site and date: Přerov, Czech Republic, 10-Apr-1985. (E) U. macrostomus, host: Cyanistes careuleus, adult male, sampling site and date: Klec, Czech Republic, 23-Jan-1967. (F) U. rossitensis, host: Sylvia atricapilla, adult male, sampling site and date: Záhlinice, Czech Republic, 7-Sep-2013.
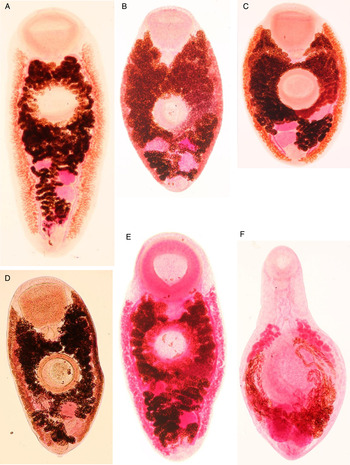
Fig. 4. Representative microphotographs of Leucochloridium holostomum (A), L. paradoxum (B), L. perturbatum (C), L. subtilis (D), L. vogtianum (E) and Leucochloridiomorpha lutea (F) stained in Semichon's carmine. (A) L. holostomum, host: Rallus aquaticus, juvenile, sampling site and date: Strachotín, Czech Republic, 23-Aug-1968. (B) L. paradoxum, host: Poecile palustris, juvenile, sampling site and date: Záhlinice, Czech Republic, 19-Jul-2014. (C) L. perturbatum, host: Vanellus vanellus, adult female, sampling site and date: Havlíčkův Brod, Czech Republic, 30-Apr-1966. (D) L. subtilis, host: Phylloscopus sibilatrix, adult male, sampling site and date: Lednice, Czech Republic, 23-Jul-1969. (E) L. vogtianum, host: Acrocephalus scirpaceus, juvenile, sampling site and date: Záhlinice, Czech Republic, 15-Aug-2013. (F) L. lutea, host: Anas platyrhynchos, juvenile, sampling site and date: Záhlinice, Czech Republic, 7-Sep-2013.
Table 6. Measurements of the three Brachylaima spp. and Urotocus rossitensis based on the adult individuals collected in the Czech Republic in years 1962–2015.

Host species of the individuals described: Brachylaima arcuatus – 30 specimens from Garrulus glandarius; B. fuscata – 30 specimens from Pica pica (5), Sturnus vulgaris (17) and Turdus merula (8); B. mesostoma – 30 specimens from Turdus merula; Urotocus rossitensis – 30 specimens from Prunella modularis (27), Turdus philomelos (2) and Sylvia borin (1). Data are shown as range (mean ± s.d.).
Table 7. Measurements of the five Leucochloridium spp., Urogonimus macrostomus, Michajlovia migrata and Leucochloridiomorpha lutea based on the adult individuals collected in the Czech Republic in years 1962–2015.
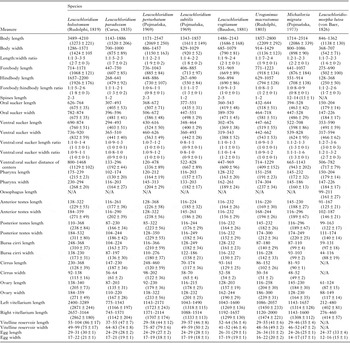
Host species of the individuals described: Leucochloridium holostomum – 30 specimens from Rallus aquaticus (25), Fulica atra (3) and Porzana porzana (2); L. paradoxum – 30 specimens from Parus major; L. perturbatum – 30 specimens from Turdus merula; L. subtilis – 30 specimens from Phylloscopus sibilatrix (5) and Turdus merula (25); L. vogtianum – 30 specimens from Acrocephalus arundinaceus; Urogonimus macrostomus – 30 specimens from Parus major; Michajlovia migrata – 9 specimens from Turdus philomelos (6), T. pilaris (2) and T. merula (1); Leucochloridiomorpha lutea – 30 specimens from Anas platyrhynchos. Data are shown as range (mean ± s.d.).
The genus Leucochloridium is characteristic by the vitellarium reaching posterior body extremity, gonads arranged in triangle, and testes in tandem. Leucochloridium vogtianum (Fig. 4E) has large body; forebody of equal length or shorter than hindbody; ventral sucker pre-equatorial; oral sucker distinctly larger than ventral; anterior testis just posterior to ventral sucker; cirrus smooth. Leucochloridium paradoxum (Fig. 4B) has round-shaped body; oral sucker slightly larger than ventral sucker; gonads close to ventral sucker. Leucochloridium perturbatum (Fig. 4C) has ventral sucker distinctly larger than oral sucker. However, L. paradoxum and L. perturbatum can be morphologically identified to species only when examining large series of individuals, the morphologic identification markers as well as the host spectrum of these two species overlap. Both these species have forebody of equal length or shorter than hindbody. Leucochloridium holostomum (Fig. 4A) has its body larger than in other leucochloridiids, with rounded extremities; ventral sucker usually larger than oral, in anterior part of body; gonads midway between ventral sucker and posterior extremity; anterior testis far from ventral sucker; cirrus postulated. Leucochloridium subtilis (Fig. 4D) has oral sucker larger than ventral sucker; uterus in intracaecal area; large bursa of Fabricius; vitellaria extend posteriorly nearly to body end. Morphologic identification of adult L. subtilis is problematic; juveniles might be confused with L. perturbatum.
The gonads arranged in triangle, testes in tandem are characteristic also for M. migrata (Fig. 3D). However, adults of this species have a large elongate body with maximum width at level of ventral sucker; suckers very large, ventral sucker larger than oral sucker, ventral sucker distant from intestinal bifurcation; vitellarium in narrow bands from pharynx to posterior margin of ovary, overlapping caeca ventrally, but not reaching caecal extremities.
The gonads arranged in line are characteristic for the genera Urogonimus and Urotocus. In U. macrostomus (Fig. 3E), vitellarium does not reach posterior body extremity, terminates at level of ovary; forebody: hindbody ratio is inconsistent. In U. rossitensis (Fig. 3F), the body is small, slender, resembling brachylaimids in shape, whole tegument spined, both suckers well developed, in anterior part of body; vitelline follicles extend either from pharynx to anterior edge of posterior testis, or from midline between suckers to posterior edge of ovary; well-developed, partially cover caeca. AdultsFootnote 3 of U. rossitensis have body oval, with maximum width at the level of ventral sucker, of similar width nearly up to the level of ovary, gradually thinning posterior from ovary up to posterior extremity; body width noticeably similar to length. In contrast to the adults, the juvenilesFootnote 4 of U. rossitensis have body elongate, with maximum width at the level of ventral sucker and anterior testis, with prominent constriction between these two sites.
The genus Leucochloridiomorpha is characteristic by the position of testes, ovary and uterus at the posterior of body, vitellaria anterior from them; body ovoid, enlarged in middle; ventral sucker much larger than oral; genital pore between or posterior to testes; uterus with two ascending and two descending limbs. The shape of testes allows distinguishing between L. lutea (Fig. 4F) and L. constantinae, with testes round or moderately oval in the first of the species and longitudinally oval in the latter species.
Another very distinct species is Amblosoma exile, which has the following identification signs: Genital pore posterior to gonads; uterus confined to region of gonads; body elongate; ventral sucker only slightly larger than oral sucker; large distance between sucker and gonads; vitelline fields short; extending only anterior to testes, distant from ventral sucker.
DISCUSSION
The identification of species of the superfamily Brachylaimoidea is not trivial, adults of some Leucochloridium spp. can be identified to species only when a large series of specimens is available, and the number of species once described, but later questioned, makes the situation especially complicated. Brachylaimoidea were first recognized by Allison (Reference Allison1943), and retained in the classifications published in the 1960s and 1970s. Phylogenetic analyses performed by Olson et al. suggested the erection of the order Diplostomida and the suborder Diplostomata, where the superfamily was classified according to the most recent classification system (Olson et al. Reference Olson, Cribb, Tkach, Bray and Littlewood2003). Branched sporocysts and cercariae with poorly developed or absent tail are typical for the Brachylaimoidea, but the life cycles differ dramatically as specified in the Introduction.
Analysis of the newly obtained DNA sequences confirmed here the classification of L. holostomum and L. vogtianum as valid species (Figs 1 and 2). Both represent type species of their subgenera (Neoleucochloridium and Papilloleucochloridium, respectively) (Gibson et al. Reference Gibson, Jones and Bray2002). However, the analysis of DNA suggested that the classification of Leucochloridium subgenera should be revised, because the DNA clades (reproducible across four independent loci) do not recapitulate the structure of subgenera suggested based on morphologic signs. The subgenus Neoleucochloridium (type species L. holostomum) is backed up by the outcomes of DNA analyses. Contrary to that, L. (Papilloleucochloridium) vogtianum forms a clade with L. (Leucochloridium) paradoxum, despite both species being considered type species of their subgenera. Contrary to the above, the third clade suggested by analyses of DNA sequences and represented by L. perturbatum and yet undescribed Leucochloridium species, is not represented by any subgenus in the current classification system of the genus Leucochloridium.
Analysis of the newly obtained DNA sequences suggested re-establishing the subfamilies Urotocinae and Urogoniminae, rejecting Urotocus and Urogonimus as members of Leucochloridiinae (Figs 1 and 2). Urotocinae were already proposed by Yamaguti (Reference Yamaguti1958) based on the differences in body shape, development of suckers and structure of sporocyst similar only to Urogonimus but not Leucochloridium (Timon-David, Reference Timon-David1957). However, Urotocinae were rejected by Gibson et al. (Reference Gibson, Jones and Bray2002), who claimed that the above features are not important at the subfamily level. The name Urogoniminae Looss, 1899 was used previously by Looss for members of the current Leucochloridiidae, but later was abandoned. Based on strong support from maximum-likelihood analyses of DNA sequences, we propose Urogonimus to be reclassified into this newly re-established subfamily.
Comparison of DNA sequences of R. capensis available in NCBI GenBank with the sequences of Brachylaima spp. led us to the rejection of Renylaima as a valid genus, suggesting to re-classify R. capensis as B. capensis (Figs 1 and 2). The genus Renylaima was described just a few years ago as a parasite of kidney and ureter of the insectivorous South-African mammal Myosorex varius (Sirgel and Mas-Coma, Reference Sirgel and Mas-Coma2010). The establishment of a new genus was based on the absence of cirrus pouch and cirrus, the presence of a genital atrium that can be evaginated to produce a prominent ventral extension of the body, existence of minor population of monotesticular form of the parasite, and parasitism in the urinary system of a mammal host (Sirgel and Mas-Coma, Reference Sirgel and Mas-Coma2010; Sirgel et al. Reference Sirgel, Artigas, Bargues and Mas-Coma2012). However, provisioning and analysis of DNA sequences of a broader spectrum of Brachylaima spp. (compared with the spectrum publicly available at a time of Renylaima description) revealed that the particular Brachylaima spp. represent rather intermediaries between R. capensis and B. arcuatus then two separate monophyletic entities, and thus the genus Renylaima should be considered synonymous to Brachylaima.
Questions still remain on both morphologic and genetical similarities of L. lutea and M. migrata. The general morphology of adults of the genus Michajlovia resembles the Leucochloridium spp., thus Pojmańska suggested the inclusion of Michajlovia in the Leucochloridiinae at the time of original description of the genus (Pojmańska, Reference Pojmańska1973). However, the position of genital pore on the ventral surface of the body and the well-developed pars prostatica suggest the classification of Michajlovia in Leucochloridiomorphidae, from which it differs in the position of the ovary, the course of the uterus and the extent of the vitellarium (Gibson et al. Reference Gibson, Jones and Bray2002). Leucochloridiomorpha lutea and M. migrata are currently recognized as members of two different families, but the sequence of their CO1 locus was similar to each other (Fig. 2A). Similarly, the position of the genital pore and the well-developed pars prostatica are shared between these two genera. They differ, however, in the position of the ovary, the course of the uterus and the extent of the vitellarium (Gibson et al. Reference Gibson, Jones and Bray2002). Elucidation of their complete life cycle of Michajlovia and more thorough DNA sequencing should provide the definitive answer on the classification of these two genera.
Concluded, the combined molecular and comparative morphological analysis of central European Brachylaimoidea led to substantial rearrangements of the systematics of Brachylaimoidea, and to the improvement on the knowledge of their host spectra, prevalence and intensity of infections. The analyses of DNA confirmed the validity of species within the genera Leucochloridium and Brachylaima, which are difficult to identify based on their morphologic characters, the validity of which was questioned previously by numerous researchers, including the authors of this manuscript.
SUPPLEMENTARY MATERIAL
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S003118201500181X
ACKNOWLEDGEMENTS
We thank Izabella Rząd (Szczecin University) for providing the helminth specimens from Poland, Teresa Pojmańska (emeritus at W. Stefański Institute of Parasitology, Warszaw) for the identification of specimens of Leucochloridium subtilis, and Milan Řezáč (Crop Research Institute, Prague) for instrumentation support.
FINANCIAL SUPPORT
The study was supported by the project PRVOUK P31/2012 from the Charles University in Prague.














