Published online by Cambridge University Press: 17 October 2003
A distinct 8 kDa calcium-binding protein (CaBP) is preferentially expressed at the cercarial stage during the life-cycle of the schistosome. Available data indicate that this CaBP may be associated with tissue/organ remodelling (involving protein degradation and synthesis of new proteins) during transformation of the cercariae from free-living form in water to parasitic life in the vertebrate host. Many CaBP molecules (e.g. calmodulin) show Ca++-dependent interaction with target proteins and thus modulate their activity. Accordingly, the parasite 8 kDa CaBP was used as a probe to clone and identify putative target protein(s) directly by binding interaction. Screening of schistosome λgt11 expression library with radio-iodinated CaBP yielded several overlapping clones showing Ca++-dependent binding of the CaBP. Sequence analyses revealed that these clones encode the S5a/Rpn10 multiubiquitin-binding protein which is a component of the regulatory 19S subunit of the 26S proteasome. The schistosome molecule, designated SmS5a, is 420 amino acids long. The nearly full length molecule (Gln3–Ser420) as well as the amino terminal (N-S5a, Gln3–Gly200) and carboxyl-terminal (C-S5a, Asp225–Ser420) portions were synthesized in bacteria, purified, and antibodies to the parasite SmS5a were prepared. Interaction between SmS5a and the 8 kDa CaBP in a Ca++-dependent manner was found under various experimental conditions: CaBP-Sepharose bound soluble SmS5a, immobilized SmS5a bound soluble CaBP, and complex formation was found when both molecules were in solution. Furthermore, it was shown that the C-terminal portion of SmS5a, but not the N-terminal portion of the molecule, reacted with the CaBP. SmS5a synthesized in a cell-free system and Western blots revealed 2 species, conceivably corresponding to the naked molecule (~50 kDa) and the molecule subjected to post-translational modification (~70 kDa). The present studies suggest that proteasome activity may be modulated by calcium, and this modulation is mediated via CaBP molecule(s).
Bilharzia is a chronic debilitating disease afflicting more than 200 million people, mainly in third world countries. The disease is caused by parasitic helminths of the genus Schistosoma, having a complex life-cycle in 2 hosts (man and snail) and short periods of larvae living freely in water (Cherfas, 1991). During the life-cycle the parasite is exposed to different environments such as fresh water at ambient temperature, and internal milieu of the hosts at 37 °C (human) or approximately 20 °C (snail). The transformation from one developmental stage to another is associated with major changes in size (0·1 to 15 mm), shape, biochemical (e.g. aerobic or anaerobic metabolism) and physiological (e.g. water tolerance or water intolerance) properties of the parasite. These changes require extensive tissue remodelling which involve protein degradation and the synthesis of new proteins. Since the 26S proteasome is the main route in eukaryotes for the degradation of intracellular proteins, it is of interest to study this system in schistosomes.
The 26S proteasome degrades short-lived and dysfunctional proteins subjected to ubiquitination. This large complex is composed of the 20S multicatalytic subunit and the 19S regulatory subunit. The 20S multicatalytic subunit confers proteolytic activity and is composed of at least 14 proteins. The 19S regulatory subunit confers ATPase activity as well as the ability to degrade ubiquitinated proteins, and it is comprised of at least 15 different proteins (Coux, Tanaka & Goldberg, 1996; Hershko & Ciehanover, 1998; Voges, Zwickl & Baumeister, 1999).
We have been studying a distinct 8 kDa calcium-binding protein (CaBP) expressed in a stage-specific manner during the life-cycle of Schistosoma mansoni. The protein is 69 amino acids long, it binds calcium and is expressed preferentially in cercariae – the larval form living freely in water (Ram et al. 1989). Immunochemical studies revealed an 8 kDa CaBP in various regions of cercariae, all of which undergo repair or reorganization upon transformation to the schistosomulum. For example, the CaBP is located in the surface tegument that within a few hours undergoes structural and physiological modifications, including doubling of the surface membrane, and changes in permeability that account for the loss of water tolerance, etc. Therefore, we proposed that the CaBP may be associated with organ/tissue remodelling during transformation from cercariae to schistosomulum, in preparation to parasitic life in the human host (Ram, Romano & Schechter, 1994). It is likely that function(s) of the 8 kDa CaBP are mediated by Ca++-dependent interactions with target proteins, thus modulating the activity of the target protein(s), similarly to calmodulin and other calcium-binding proteins (Klee & Vanaman, 1982; Da Silva & Reinach, 1991). To identify putative target protein(s), a cDNA expression library prepared from mRNA of adult worms of S. mansoni, was screened with 125I-labelled 8 kDa CaBP as a probe. A few clones which interacted with the CaBP in a Ca++-dependent manner were sequenced and identified as the S5a/Rpn10 multiubiquitin chain-binding protein which is a component of the 19S regulatory subunit of the 26S proteasome (Ferrell et al. 1996; van Nocker et al. 1996 a). The parasite protein was named SmS5a (Schistosoma mansoni homologue of the S5a/Rpn10 protein).
The Puerto Rican strain of S. mansoni was maintained by passage through Biomphalaria glabrata snails and ICR mice. Parasites from different developmental stages of the life-cycle, and protein lysates of the parasite were prepared as described (Ram et al. 1994).
The cercariae-specific 8 kDa CaBP was synthesized in bacteria using the pET expression vector (Studier et al. 1990). The CL17 cDNA clone encoding the full length CaBP (Ram et al. 1989) served as template for PCR using appropriate primers with built-in restriction sites for cloning into the pET plasmid. The protein was isolated from transformed bacteria induced by isopropyl β-D-thiogalactopyranoside (50 mg CaBP/l of bacterial culture). The bacterial lysate was loaded on DE52 anion-exchanger (0·05 M Tris–HCl, pH 7·3) and eluted using an NaCl gradient (0·1–0·4 M). SDS–PAGE showed a single band of CaBP (>98% pure). The purity of the protein was estimated by running serial dilutions of the protein preparation on SDS–PAGE, staining the gel with Coomassie blue, estimating intensities of the protein bands, and comparison of the non-relevant band(s) to the band of the CaBP (or CaBP-Tyr).
A tyrosine codon was added to the 3′ end of the coding region of the CaBP CL17 cDNA clone by PCR (Weiner et al. 1994). The modified cDNA was expressed in the pET vector to yield CaBP-Tyr in which 1 Tyr residue was added to the C-terminus of the 8 kDa CaBP. The CaBP-Tyr was isolated from bacterial lysate by NaCl gradient elution from DE52 anion-exchanger as described above. The CaBP-Tyr (>92% pure by SDS–PAGE) was 125I-labelled by the chloramine-T method (McConahey & Dixon, 1980).
The PTP-7 cDNA encoding nearly full length SmS5a (Ser2–Ser420) in Bluescript KS, served as template for PCR using primers with built in restriction sites for cloning into the pGEX (Smith & Johnson, 1988) or pJC40 (Clos & Brandau, 1994) expression vectors. Nearly complete SmS5a (Gln3–Ser420), as well as the N-terminal fragment (N-S5a, Gln3–Gly200) and the C-terminal fragment (C-S5a, Asp225–Ser420) were synthesized using the pGEX vector to yield fusion proteins consisting of SmS5a moieties coupled to GST (glutathione S-transferase of S. japonicum) that were affinity purified on GSH-agarose (Smith & Johnson, 1988). Another C-terminal fragment (Asp225–Ser420) with His-tail was synthesized using the pJC40 vector, and purified by Ni-resin chromatography (Clos & Brandau, 1994).
Recombinant fusion proteins of N-S5a or C-S5a coupled to GST were injected into rabbits (100 μg per injection) at 12-day intervals as described (Neuman et al. 1993). Antisera were collected 12 days after the third injection. Four affinity columns were prepared by coupling proteins to Sepharose-4B activated by the cyanogen bromide procedure (Axen, Porath & Ernback, 1967): C-S5a-GST (3 mg protein/g wet gel), C-S5a-His (5 mg protein/g wet gel), GST (4 mg protein/g wet gel), lysate of E. coli HB101 bacteria (6 mg protein/g wet gel). Antisera were first passed on Sepharose–bacterial lysate conjugate. The flow-through was passed on a second column of Sepharose-GST conjugate. The flow-through was passed on a third column of Sepharose-C-S5a-GST or Sepharose-C-S5a-His. After washing the column with phosphate-buffered saline, pH 7·2 (PBS), the adsorbed antibodies were eluted with 0·1 M acetic acid, neutralized with Tris base to pH 7·2 and dialysed against PBS at 4 °C.
Total RNA of mouse kidney, hepatopancreas (HP) of snails, and of S. mansoni at various developmental stages, were prepared according to Kirby (1968). DNA nucleotide sequences were determined at least 3 times on overlapping segments of different clones and on both strands by means of an automated sequencer (ABI DNA sequencer, model 373A). Other procedures, including RNA translation in the wheat germ extract and immunoprecipitation of the cell-free products (Parvari et al. 1987), SDS–PAGE, Western blots developed with rabbit antibodies and 125I-labelled purified goat antibodies to rabbit immunoglobulins, and Northern blots in formaldehyde gels, were done according to published procedures (Sambrook, Fritsch & Maniatis, 1989).
Agar plates with plaques of λgt11 expression library of schistosome cDNA (10000 PFU per 15 cm plate) were overlayed with a nitrocellulose filter for 5 min. The filters were washed for 15 min in binding buffer (50 mM Tris–HCl, pH 7·4, 150 mM NaCl) containing 1 mM CaCl2 and 10% (v/v) skim milk. The filters were kept overnight at room temperature in 150 ml of the same solution to which 80 μg of 125I-labelled CaBP-Tyr was added (3·6×106 cpm/ml). Subsequently, filters were washed 3 times (5 min each) with binding buffer containing 0·05% Tween 20, dried and exposed to X-ray film. The radioactive binding solution was kept at 4 °C for further use with sodium azide (0·03%) (Fromm & Chua, 1992).
Parasite and bacterial lysates, as well as recombinant SmS5a proteins, were subjected to SDS–PAGE, proteins were electrotransfered to a nitrocellulose filter, reacted with 125I-CaBP-Tyr together with CaCl2 (1 mM) or EGTA (10 mM), washed and exposed to X-ray film, as described above for screening the cDNA library.
The CaBP was coupled to CNBr-activated Sepharose-4B to yield a conjugate containing 5·5 mg CaBP/ml wet gel. CaBP-Sepharose (50 μl wet gel containing 275 μg of immobilized CaBP) and SmS5a or fragments of SmS5a (125 μg protein) were mixed in a solution containing 50 mM Tris–HCl, pH 7·4, 1 mM DTT and either 1 mM CaCl2 or 10 mM EGTA. The reaction mixture (200 μl) was kept for 1 h at room temperature. Sepharose beads were precipitated by centrifugation and washed twice with 1 ml of Tris–HCl, pH 7·4 buffer containing 1 mM CaCl2 or 10 mM EGTA. Bound proteins were eluted from the beads by incubation at room temperature for 30 min in a solution (100 μl) containing 20 mM EGTA and 50 mM Tris–HCl, pH 7·4. Aliquots from the reaction mixture, washing solution, and eluate were analysed by SDS–PAGE.
The CaBP was used as an affinity probe to identify cDNA clones encoding the putative target protein(s). To this aim we added 1 Tyr codon to the molecule (the 8 kDa CaBP has no tyrosine residues) and prepared CaBP-Tyr that can be labelled with radioactive iodine. The recombinant CaBP (>98% pure) and CaBP-Tyr (>92% pure) molecules had similar mobilities on SDS–PAGE (apparent Mr of 11000, see Ram et al. 1989). Authenticity of the CaBP was ascertained by sequence analysis, showing amino terminal sequence (12 residues) identical to the sequence predicted from the nucleotide sequence of the CL17 cDNA. The CaBP and CaBP-Tyr bound 45Ca++ (Maruyama, Mikawa & Ebashi, 1984), and showed Ca++-dependent electrophoretic mobility shifts similar to other CaBP molecules (Klee, Crouch & Krinks, 1972). Both molecules had increased mobility with CaCl2 and decreased mobility with EGTA (data not shown).
Lysates of cercariae and adult worm resolved on SDS–PAGE were overlaid with 125I-CaBP-Tyr. Both lysates revealed 2 bands of about 50 kDa and 30 kDa reacting with 125I-CaBP-Tyr in the presence of CaCl2 (1 mM) but not with EGTA (10 mM). Based on these findings (data not shown) we decided to screen the adult worm cDNA library for putative target protein(s), because this library (kindly donated by Dr A. J. G. Simpson) had longer inserts than our cercarial cDNA libraryl. A λgt11 cDNA library of adult worms of S. mansoni (280000 PFU) was screened with 125I-CaBP-Tyr and 1 mM CaCl2. Sixteen independent clones were purified, all of which bound 125I-CaBP-Tyr in the presence of CaCl2 but not with EGTA (10 positive clones are shown in Fig. 1).

Fig. 1. Calcium-dependent interaction of λgt11 clones encoding SmS5a with 125I-CaBP-Tyr. Purified phages were spotted onto a lawn of E. coli, plaques were transferred to a nitrocellulose filter and reacted with 125I-CaBP-Tyr probe in the presence of 1 mM CaCl2 (A) or 10 mM EGTA (B). Asterisks indicate 1 unrelated plaque showing weak non-specific binding of the probe in the presence of CaCl2 or EGTA.
The cDNA inserts (0·6–1·6 kb) of the 16 positive λgt11 clones were subcloned into Bluescript KS and sequenced. Alignment of the sequences showed overlapping segments, all of which were included in the longest PTP-7 clone of 1578 bp, starting with Ser2 codon and ending with polyA tail of 32 adenines. A Met codon (designated Met+1, see below) preceding Ser2 and additional 9 codons were deduced from SmS5a genomic clone in which the promoter and adjacent coding region were sequenced (Accession no. AF502283). It is likely that Met+1 is the initiator methionine since it aligns with the initiator-Met of S5a/Rpn10 proteins of other species (see Fig. 2), in the genomic clone an in-frame termination codon was found at position −10 and there was no Met codon between it and the Met+1 codon (unpublished data). Accordingly, the open reading frame, starting from the Met+1 codon and ending with a UAA termination codon, is 1260 bases long encoding 420 amino acid, and the 3′ untranslated region is 286 bases long plus 32 adenines of the polyA tail. Sequence comparisons to the GenBank database revealed that PTP-7 encodes the parasite homologue of the S5a/Rnp10 protein which is part of the 19S regulatory subunit of the 26S proteasome. The parasite protein, designated SmS5a, is the longest S5a/Rpn10 molecule known so far (Fig. 2). Harrop, Coulson & Wilson (1999) cloned the S5a of S. mansoni (clone NLSL19, Accession no. AF030960) by screening parasite expression cDNA library with antiserum raised against proteins released by the lung-stage parasite. The deduced amino acid sequence of the NLSL19 cDNA is 100% identical to the SmS5a isolated in our laboratory. Nucleotide sequences are also identical, except for the 3′ untranslated region where PTP-7 has an additional 137 bases, probably due to the usage of a different poly-adenylation addition site. In both clones the typical AATAAA poly-adenylation signal is missing.

Fig. 2. Comparison of SmS5a with the amino acid sequences of S5a/Rpn10 from other species. The Wisconsin Package Version 10.2 (Genetics Computer Group, Madison, Wisconsin, Tel.: (608) 231 5200, E-mail: help@gcg.com) was used to align the sequence of schistosome SmS5a (predicted from the nucleotide sequence of the cDNA) with amino acid sequences of S5a/Rpn10 of human (Ferrell et al. 1996), D. melanogaster (Haracska & Udvary, 1995), A. thaliana (van Nocker et al. 1996 a) and yeast (van Nocker et al. 1996 b). Identical residues are denoted by white letters on black. Asterisks indicate the beginning and end of recombinant SmS5a moieties (SmS5a-GST:Gln3-Ser420, N-S5a-GST:Gln3-Gly200, C-S5a-GST:Asp225-Ser420, C-S5a-His:Asp225-Ser420). Circles mark the hydrophobic cores of PUbS1 and PUbS2 (Young et al. 1998). The segment of SmS5a (Leu345-Leu365) proposed to dock the 8 kDa CaBP is lined above. A short segment (17 or 19 amino acids) highly enriched with alanine or glycine residues in A. thaliana and yeast proteins is marked by tidals. Nucleotide sequences of the SmS5a cDNA from the Met1 codon to the polyA tail (Accession no. AF502282) and of the genomic clone of SmS5a covering 335 nucleotides of the promoter region till codon 152 of the cDNA (Accession no. AF502283) have been submitted to the GenBank™ database. Potential sites for phosphorylation and glycosylation of SmS5a can be found in the cDNA sequence deposited in GenBank.
The specificity of interaction between SmS5a and 125I-CaBP-Tyr was ascertained by competition experiments using cold CaBP-Tyr and CaBP as inhibitors. Lysates of bacteria expressing C-S5a-GST (C-S5a moiety of SmS5a that binds CaBP, see below) were reacted with 125I-CaBP-Tyr either alone, or with added unlabelled CaBP or CaBP-Tyr. As seen in Fig. 3 the unlabelled proteins competed for the binding of 125I-CaBP-Tyr. These results demonstrate that addition of tyrosine or iodo-tyrosine to CaBP did not significantly perturb interaction with the SmS5a molecule.

Fig. 3. Specificity of interaction between SmS5a and 125I-CaBP-Tyr. Decreasing amounts of bacterial lysate expressing C-S5a-GST were loaded on SDS–PAGE: 1 μl lysate in lane 1, and 1 μl of 2-fold dilutions in adjacent lanes (1[ratio ]2 in lane 2 to 1[ratio ]32 in lane 6). After electrophoresis proteins were transferred to a nitrocellulose filter and reacted with 125I-CaBP-Tyr alone (A), with added 8 kDa CaBP (16 μg/ml) (B), or CaBP-Tyr (16 μg/ml) (C).
The predicted amino-acid sequence of SmS5a revealed that the protein can be divided into 2 domains, a neutral N-terminal portion (residues 3–200, calculated pI 7·47) designated N-S5a, and an acidic C-terminal portion (residues 225–420, calculated pI 3·95) designated C-S5a. The nearly full length SmS5a (residues 3–420) as well as the N-terminal and C-terminal fragments were expressed in bacteria as fusion proteins with GST, designated, respectively, SmS5a-GST, N-S5a-GST and C-S5a-GST. The C-terminal portion (residues 225–420) was also expressed with a short tail of 10 histidine residues to yield the C-S5a-His fragment. Recombinant proteins isolated from the bacterial lysates (see Materials and Methods section) analysed on SDS–PAGE showed a major band of the purified recombinant protein. Contamination by unrelated proteins was small (<5%) for GST, C-S5a-GST and C-S5a-His. A minor doublet of proteins of ~30 kDa was found in the N-S5a-GST and SmS5a-GST protein preparations (Fig. 4A). The ~30 kDa proteins conceivably represent GST plus a few amino acids from the N-terminus of SmS5a, generated by premature termination of mRNA translation (Schechter et al. 1975).

Fig. 4. Ca++-dependent interaction of the 8 kDa CaBP with SmS5a and derivatives immobilized on nitrocellulose filter. (A) Coomassie blue staining of SDS–PAGE of purified GST (1), C-S5a-GST (2), N-S5a-GST (3), SmS5a-GST (4) and C-S5a-His (5). Autoradiograms of SDS–PAGE of gels identical to (A) after transfer to nitrocellulose filter and overlay with 125I-CaBP-Tyr in binding buffer containing 1 mM CaCl2 (B) or 10 mM EGTA (C). Scheme of SmS5a protein fragments synthesized in bacteria is given at the top.
SmS5a and derivatives were electrophoresed on SDS–PAGE, blotted and overlaid with 125I-CaBP-Tyr. The SmS5a-GST, C-S5a-GST and C-S5a-His showed binding of 125I-CaBP-Tyr in the presence of CaCl2 but not in the presence of EGTA, while N-S5a-GST did not bind 125I-CaBP-Tyr under any conditions. GST, serving as control, was also negative for 125I-CaBP-Tyr binding in the presence of CaCl2 or EGTA (Fig. 4). These findings demonstrate Ca++-dependent binding of CaBP by SmS5a, and map the binding activity to the C-terminal portion of the molecule. Binding experiments quoted below using CaBP-Sepharose and SmS5a-GST fusion proteins immobilized on GSH-agarose, confirm these findings.
SmS5a and derivatives were mixed with CaBP-Sepharose beads. In the presence of 1 mM CaCl2 the SmS5a-GST (data not shown), C-S5a-GST (data not shown) and C-S5a-His (Fig. 5) were retained on the beads, and retained proteins were released by washing with 20 mM EGTA. In the presence of 10 mM EGTA no protein was retained on the CaBP-Sepharose (Fig. 5). The N-S5a-GST and GST proteins were not bound to the CaBP-Sepharose in the presence of CaCl2 or EGTA (data not shown).

Fig. 5. Ca++-dependent interaction of C-S5a-His with CaBP-Sepharose. C-S5a-His (125 μg) was reacted with CaBP-Sepharose (275 μg immobilized 8 kDa CaBP) in the presence of CaCl2 (1 mM) or EGTA (10 mM). (A) Sepharose beads mixed with C-S5a-His (1) and washed (2) in the presence of 1 mM CaCl2. (B) Sepharose beads mixed with C-S5a-His (4) and washed (5) in the presence of 10 mM EGTA. Beads were then incubated in 20 mM EGTA (3, 6). Aliquots from the supernatant of the mixture of beads with C-S5a-His (1 and 4), from washing of the beads (2 and 5), and eluates from beads treated with 20 mM EGTA (3 and 6) were resolved on SDS–PAGE and stained with Coomassie blue.
Similar results were obtained when the SmS5a and derivatives were immobilized and the CaBP was in solution. The SmS5a-GST, C-S5a-GST and N-S5a-GST were immobilized by adsorption to GSH-agarose beads (Sigma), and reacted with soluble CaBP in the presence of 1 mM CaCl2 or 10 mM EGTA. The beads were washed and any adsorbed CaBP was released by boiling in gel-loading buffer, and the CaBP was detected by Western blot (Fig. 6). In agreement with previous experiments (Figs 4 and 5), the nearly full length protein (SmS5a-GST) and the C-terminal fragment (C-S5a-GST) bound CaBP in a Ca++-dependent manner, whereas the N-terminal fragment (N-S5a-GST) and GST did not bind CaBP. In this system we tested possible interactions between bovine calmodulin (Sigma) with immobilized Sm5a-GST and derivatives. The calmodulin was not bound by SmS5a-GST or C-S5a-GST under conditions in which the parasite 8 kDa CaBP was bound, and it was not bound by N-S5a-GST (data not shown).

Fig. 6. Ca++-dependent interactions of the 8 kDa CaBP with SmS5a and derivatives immobilized on GSH-agarose. GST-fusion proteins (~20 μg) were adsorbed on GSH-agarose beads (20 μl wet gel in 200 μl) according to instructions of the manufacturer (Sigma): SmS5a-GST (B), C-S5a-GST (C), N-S5a-GST (D) and GST (E). GSH-Agarose without added protein (A). Beads were equilibrated with 1 mM CaCl2 (1) or 5 mM EGTA (4), mixed with 8 kDa CaBP (2 μg protein) in 25 mM Hepes, pH 7·6, 100 mM NaCl, 10 mM DTT containing 1 mM CaCl2 (2) or 5 mM EGTA (5), washed in same solution, and adsorbed CaBP was extracted from the beads by boiling in gel loading buffer (3, 6). Aliquots from the equilibration solutions (1, 4), from the supernate of beads mixed with CaBP (2, 5) and from the gel loading buffer (3, 6) were resolved on SDS–PAGE, and subjected to Western blot using rabbit purified antibodies to the CaBP and 125I-labelled goat purified antibodies to rabbit IgG.
To check whether SmS5a forms complexes with the CaBP in solution, 7 μg of C-S5a-GST were mixed with 3·6 μg of CaBP in the presence of either 1 mM CaCl2 or 5 mM EGTA. After 1 h at room temperature purified antibodies against C-S5a-GST were added, the immune precipitate was dissolved in gel-loading buffer and resolved on SDS–PAGE, blotted, and the Western blot was developed with purified antibodies against the CaBP. CaBP was detected only when CaCl2 was present in the reaction (data not shown). These results demonstrate Ca++-dependent complex formation between CaBP and C-S5a-GST in solution.
To evaluate whether transcription of the SmS5a gene is regulated during the life-cycle of the parasite, we analysed the RNAs of miracidia, sporocysts, cercariae, and adult worms. Northern blot showed a single SmS5a mRNA band of 1·6 kb, in agreement with the size of the cDNA. The mRNA was found in all life-stages tested, yet mRNA levels were variable and were highest in adult worms. The control RNAs, from hepatopancreas of uninfected snails and mouse kidney, did not show any hybridizing band (Fig. 7).

Fig. 7. Transcription of the SmS5a mRNA during the life-cycle of Schistosoma mansoni. Northern blot probed with 32P-labelled PTP-7 cDNA encoding SmS5a. Total RNA (10 μg/lane) was extracted from mouse kidney (MK), hepatopancreas (HP) of uninfected snails, miracidia (MIR), hepatopancreas of infected snails containing sporocysts (SPO), cercariae (CER), and adult worms (AW). The position of the 18S rRNA is indicated.
The RNA from adult worms was translated in the wheat germ cell-free system, and the 35S-Met labelled cell-free products were immunoprecipitated by different antibodies. Antisera to N-S5a-GST (data not shown) and C-S5a-GST (Fig. 8, lane 3) precipitated an ~50 kDa protein similar to the mass of the full-length SmS5a protein (45·7 kDa) predicted from the cDNA sequence. As expected, antisera to the GST-fusion proteins and antiserum to GST precipitated the 28 kDa glutathione S-transferase of the parasite (Fig. 8, lanes 2, 3). When purified antibodies to the C-S5a moiety were prepared (adsorption of antiserum to C-S5a-GST on 3 immunoadsorbents to remove anti-GST antibodies, see Materials and Methods section) only the ~50 kDa band was precipitated (Fig. 8, lane 6). Controls of pre-immune sera and of purified antibodies to BSA did not precipitate any protein (Fig. 8, lane 1, 4). Antisera to N-S5a-GST and C-S5a-GST did not precipitate any protein from cell-free products programmed by mouse kidney RNA in the wheat germ extract (data not shown).

Fig. 8. SDS–PAGE of immunoprecipitated 35S-Met labelled proteins programmed by mRNA of Schistosoma mansoni using different antibodies. Cell-free products synthesized in the wheat germ extract were programmed by RNA extracted from adult worms. The same amount of protein product (500000 cpm of TCA precipitable material) was reacted with rabbit pre-immune serum (1) and with rabbit antisera against GST (2) or C-S5a-GST (3), as well as with purified antibodies against bovine serum albumin (4), GST (5), and C-S5a moiety of C-S5a-GST (see Materials and Methods section). The positions of protein size markers (kDa) are indicated.
Western blots of protein lysates from mouse kidney, cercariae and adult worm, were developed with purified antibodies against BSA, GST and the C-S5a moiety. Anti-BSA, which served as a control, did not interact with any protein of the lysates (Fig. 9A). Anti-GST (elicited against S. japonicum GST) revealed in S. mansoni lysates 2 bands of 28 kDa and 35 kDa, representing immunological cross-reaction with S. mansoni GST's, and it did not interact with the mouse kidney lysate. Anti-C-S5a revealed in parasite lysates 2 proteins of about 50 kDa and 70 kDa. The 50 kDa protein is likely to be the ‘naked’ SmS5a, while the 70 kDa protein is probably SmS5a subjected to post-translational modification (see Discussion section). In cercariae the putative naked SmS5a protein (50 kDa) was more abundant than the modified SmS5a (70 kDa), while in adult worms the 70 kDa protein was more abundant than the 50 kDa protein. The 70 kDa species was more abundant in adult worms than in cercariae. Two minor proteins of about 40 kDa and 36 kDa, present in adult worms but not in cercariae, may represent fragments of SmS5a.

Fig. 9. Expression of the SmS5a protein in cercariae and adult worms. Lysates (40 μg protein/lane) were prepared from mouse kidney (1), cercariae (2) and adult worms (3). Western blots were reacted with purified antibodies to BSA (A), GST (B), and C-S5a moiety of C-S5a-GST (C), and then with 125I-labelled goat purified antibodies to rabbit IgG. The positions of protein size markers (kDa) are indicated.
Many metabolic (e.g. activation of kinases) and physiological (e.g. muscle contraction) events triggered by Ca++ are mediated via CaBP molecules that interact with target proteins and modulate their activity (Klee, 1982; Da Silva & Reinach, 1991). Therefore it was of interest to identify target proteins interacting with the 8 kDa CaBP preferentially expressed at the cercarial stage during the life-cycle of schistosomes. These studies would clarify which function specific to cercariae the 8 kDa CaBP performs. By using the 8 kDa CaBP as a protein probe we cloned from the cDNA expression library of the parasite the SmS5a molecule corresponding to the S5a/Rpn10 multiubiquitin-binding protein. Interaction of the 8 kDa CaBP with the SmS5a component of the 26S proteasome indicates that it may be involved with protein degradation. These findings support an earlier proposal, based on immunohistochemical studies (Ram et al. 1994), that the 8 kDa CaBP is associated with tissue/organ remodelling of cercariae in preparation for parasitic life in the vertebrate host.
The mRNA and protein of SmS5a were found in cercariae in which the 8 kDa CaBP mRNA and protein are expressed, as well as in adult worms in which the CaBP mRNA and protein are undetectable (Ram et al. 1989, 1994). The SmS5a may gain cercariae-specific function by interaction with the cercariae-specific 8 kDa CaBP. In adult worms SmS5a may function differently either as a free molecule, or due to complex formation with another CaBP that may confer different properties on SmS5a, say, facilitation (or inhibition) of interaction with different sets of multiubiquitinated proteins.
Western blots revealed 2 bands reacting with purified antibodies to SmS5a. The 50 kDa band is likely to be the unmodified protein since it is similar in size to the SmS5a synthesized in the cell-free system in which post-translation modifications do not occur. The 70 kDa species may be a modified SmS5a that contains concensus sequences for glycosylation and phosphorylation (see legend to Fig. 2). Early experiments strongly suggested that S5a/Rpn10 of Drosophila is glycosylated (Haracska & Udvary, 1995).
The Northern and Western blots show that the levels of SmS5a mRNA and protein, as well as the ratio of modified and unmodified SmS5a protein species, differ at different developmental stages, demonstrating regulation of SmS5a expression during the life-cycle of the parasite. These findings are consistent with reports on other organisms in which the expression of proteasome components varies among tissues and at different stages of development (Coux et al. 1996; Voges et al. 1999).
We have shown that SmS5a interacts with the cercarial specific 8 kDa CaBP in a Ca++-dependent manner under various experimental conditions. It was found that C-S5a-GST, that binds the parasite 8 kDa CaBP, did not bind bovine calmodulin. Further studies are needed to clarify if this reflects species specificity of the reactants, or whether SmS5a binds the cercarial specific 8 kDa CaBP but not other CaBP molecules of the parasite.
The predicted amino acid sequence of SmS5a revealed that the protein could be divided into a neutral N-terminal portion (pI 7·47) and an acidic C-terminal portion (pI 3·93), that were synthesized in bacteria. Binding experiments showed that the C-terminal portion of SmS5a, but not the N-terminal portion of the molecule, reacted with the CaBP in a Ca++-dependent manner.
Alignments of S5a/Rpn10 from different organisms (mammals, flies, yeast, plants) demonstrate that the amino acid sequence of the N-terminal portion of the molecule is well conserved, more than the C-terminal portion. The function of the N-terminal portion is not yet known. Proteolytic mapping and deletion analyses of S5a/Rpn10 have shown that the C-terminal portion binds multiubiquitin chains (Ferrell et al. 1996; van Nocker et al. 1996 a,b), and 2 independent binding sites were mapped: PUbS1 and PUbS2 (Young et al. 1998). The multiubiquitin binding sites are approximately 30 amino acids long and are separated by about 50 residues. The hydrophobic cores of the PUbS1 and PUbS2 binding sites are fairly conserved in the parasite SmS5a. The schistosome SmS5a is longer than S5a/Rpn10 of other species, and it has an additional segment apparently not found in other S5a/Rpn10 molecules. The putative unique segment is about 21 amino acids long (positions 345–365), it is enriched with amino acids carrying side-chains with hydroxyl groups (6 Ser and 3 Thr residues) and with amino acids with short side-chains (5 Ala, Gly, Pro). We tentatively propose this segment as the candidate to dock the 8 kDa CaBP. It is seen that A. thaliana and yeast S5a/Rpn10 proteins also contain unique short segments (17 or 19 amino acids long) highly enriched with alanine or glycine that potentially may function like the above 21 amino acid segment.
The activity of the proteasome is regulated by various mechanisns that affect specificity and efficacy of peptide degradation. For example, interferon-γ (IFN-γ) induces the replacement of 3 β subunits by LMP subunits which facilitate antigen presentation. The LMP subunits modify cleavage-site preference of the 26S proteosome, to yield peptides favourable for presentation on MHC class I molecules (Goldberg & Rock, 1992; Pamer & Cresswell, 1998). IFN-γ also causes increased levels of the PA28 regulatory molecule that can associate with the 20S subunit (Realini et al. 1994; Ahn et al. 1995). This association markedly increases the hydrolysis of peptides, without affecting the rate of hydrolysis of proteins. PA28 is composed of 2 subunits, one of which (P28α) is a 28 kDa protein with KEKE – motif (Lys, Glu repeats) that binds calcium ions. Calcium can reversibly inhibit the activation of proteasome's peptidase activity induced by PA28 (Realini & Rechsteiner, 1995). Involvement of calcium in proteasome activity was reported during ascidian meiotic division cycle (Kawahara & Yokosawa, 1994) and during Xenopus egg activation (Aizawa et al. 1996). It was proposed that proteasome activation towards ubiquitinated proteins (e.g. cyclin) is due to assembly of the 26S proteasome from the 20S subunit, and this interconversion is regulated by intracellular calcium release. These experiments raise the possibility that proteasome activity is regulated by calcium. Yet, the physiological relevance and mechanism of proteasome regulation by calcium are not clear (Realini & Rechsteiner, 1995; Kawahara & Yokosawa, 1994; Aizawa et al. 1996).
The present studies demonstrating Ca++-dependent interaction between the schistosome SmS5a and the 8 kDa CaBP, indicate that proteasome function can be modulated by calcuim, but this effect is mediated via calcium binding protein molecules. Further experiments are needed to evaluate the significance of calcium and CaBP interaction with S5a/Rnp10 on proteasome function, and effect of this interaction on organ/tissue remodeling during the life-cycle of schistosome.
We thank Dr Andrew J. G. Simpson (Instituto Ludwig de Pesquisa sobre o Cancer, Sao Paulo) for providing the λgt11 cDNA library of S. mansoni. This work was supported in part by grants from the UNPD/World Bank/WHO Special Program for Research and Training in Tropical Diseases and the Israel Science Foundation.
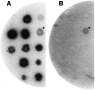
Fig. 1. Calcium-dependent interaction of λgt11 clones encoding SmS5a with 125I-CaBP-Tyr. Purified phages were spotted onto a lawn of E. coli, plaques were transferred to a nitrocellulose filter and reacted with 125I-CaBP-Tyr probe in the presence of 1 mM CaCl2 (A) or 10 mM EGTA (B). Asterisks indicate 1 unrelated plaque showing weak non-specific binding of the probe in the presence of CaCl2 or EGTA.

Fig. 2. Comparison of SmS5a with the amino acid sequences of S5a/Rpn10 from other species. The Wisconsin Package Version 10.2 (Genetics Computer Group, Madison, Wisconsin, Tel.: (608) 231 5200, E-mail: help@gcg.com) was used to align the sequence of schistosome SmS5a (predicted from the nucleotide sequence of the cDNA) with amino acid sequences of S5a/Rpn10 of human (Ferrell et al. 1996), D. melanogaster (Haracska & Udvary, 1995), A. thaliana (van Nocker et al. 1996 a) and yeast (van Nocker et al. 1996 b). Identical residues are denoted by white letters on black. Asterisks indicate the beginning and end of recombinant SmS5a moieties (SmS5a-GST:Gln3-Ser420, N-S5a-GST:Gln3-Gly200, C-S5a-GST:Asp225-Ser420, C-S5a-His:Asp225-Ser420). Circles mark the hydrophobic cores of PUbS1 and PUbS2 (Young et al. 1998). The segment of SmS5a (Leu345-Leu365) proposed to dock the 8 kDa CaBP is lined above. A short segment (17 or 19 amino acids) highly enriched with alanine or glycine residues in A. thaliana and yeast proteins is marked by tidals. Nucleotide sequences of the SmS5a cDNA from the Met1 codon to the polyA tail (Accession no. AF502282) and of the genomic clone of SmS5a covering 335 nucleotides of the promoter region till codon 152 of the cDNA (Accession no. AF502283) have been submitted to the GenBank™ database. Potential sites for phosphorylation and glycosylation of SmS5a can be found in the cDNA sequence deposited in GenBank.
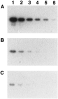
Fig. 3. Specificity of interaction between SmS5a and 125I-CaBP-Tyr. Decreasing amounts of bacterial lysate expressing C-S5a-GST were loaded on SDS–PAGE: 1 μl lysate in lane 1, and 1 μl of 2-fold dilutions in adjacent lanes (1[ratio ]2 in lane 2 to 1[ratio ]32 in lane 6). After electrophoresis proteins were transferred to a nitrocellulose filter and reacted with 125I-CaBP-Tyr alone (A), with added 8 kDa CaBP (16 μg/ml) (B), or CaBP-Tyr (16 μg/ml) (C).
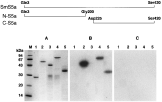
Fig. 4. Ca++-dependent interaction of the 8 kDa CaBP with SmS5a and derivatives immobilized on nitrocellulose filter. (A) Coomassie blue staining of SDS–PAGE of purified GST (1), C-S5a-GST (2), N-S5a-GST (3), SmS5a-GST (4) and C-S5a-His (5). Autoradiograms of SDS–PAGE of gels identical to (A) after transfer to nitrocellulose filter and overlay with 125I-CaBP-Tyr in binding buffer containing 1 mM CaCl2 (B) or 10 mM EGTA (C). Scheme of SmS5a protein fragments synthesized in bacteria is given at the top.

Fig. 5. Ca++-dependent interaction of C-S5a-His with CaBP-Sepharose. C-S5a-His (125 μg) was reacted with CaBP-Sepharose (275 μg immobilized 8 kDa CaBP) in the presence of CaCl2 (1 mM) or EGTA (10 mM). (A) Sepharose beads mixed with C-S5a-His (1) and washed (2) in the presence of 1 mM CaCl2. (B) Sepharose beads mixed with C-S5a-His (4) and washed (5) in the presence of 10 mM EGTA. Beads were then incubated in 20 mM EGTA (3, 6). Aliquots from the supernatant of the mixture of beads with C-S5a-His (1 and 4), from washing of the beads (2 and 5), and eluates from beads treated with 20 mM EGTA (3 and 6) were resolved on SDS–PAGE and stained with Coomassie blue.

Fig. 6. Ca++-dependent interactions of the 8 kDa CaBP with SmS5a and derivatives immobilized on GSH-agarose. GST-fusion proteins (~20 μg) were adsorbed on GSH-agarose beads (20 μl wet gel in 200 μl) according to instructions of the manufacturer (Sigma): SmS5a-GST (B), C-S5a-GST (C), N-S5a-GST (D) and GST (E). GSH-Agarose without added protein (A). Beads were equilibrated with 1 mM CaCl2 (1) or 5 mM EGTA (4), mixed with 8 kDa CaBP (2 μg protein) in 25 mM Hepes, pH 7·6, 100 mM NaCl, 10 mM DTT containing 1 mM CaCl2 (2) or 5 mM EGTA (5), washed in same solution, and adsorbed CaBP was extracted from the beads by boiling in gel loading buffer (3, 6). Aliquots from the equilibration solutions (1, 4), from the supernate of beads mixed with CaBP (2, 5) and from the gel loading buffer (3, 6) were resolved on SDS–PAGE, and subjected to Western blot using rabbit purified antibodies to the CaBP and 125I-labelled goat purified antibodies to rabbit IgG.
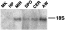
Fig. 7. Transcription of the SmS5a mRNA during the life-cycle of Schistosoma mansoni. Northern blot probed with 32P-labelled PTP-7 cDNA encoding SmS5a. Total RNA (10 μg/lane) was extracted from mouse kidney (MK), hepatopancreas (HP) of uninfected snails, miracidia (MIR), hepatopancreas of infected snails containing sporocysts (SPO), cercariae (CER), and adult worms (AW). The position of the 18S rRNA is indicated.
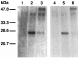
Fig. 8. SDS–PAGE of immunoprecipitated 35S-Met labelled proteins programmed by mRNA of Schistosoma mansoni using different antibodies. Cell-free products synthesized in the wheat germ extract were programmed by RNA extracted from adult worms. The same amount of protein product (500000 cpm of TCA precipitable material) was reacted with rabbit pre-immune serum (1) and with rabbit antisera against GST (2) or C-S5a-GST (3), as well as with purified antibodies against bovine serum albumin (4), GST (5), and C-S5a moiety of C-S5a-GST (see Materials and Methods section). The positions of protein size markers (kDa) are indicated.
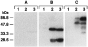
Fig. 9. Expression of the SmS5a protein in cercariae and adult worms. Lysates (40 μg protein/lane) were prepared from mouse kidney (1), cercariae (2) and adult worms (3). Western blots were reacted with purified antibodies to BSA (A), GST (B), and C-S5a moiety of C-S5a-GST (C), and then with 125I-labelled goat purified antibodies to rabbit IgG. The positions of protein size markers (kDa) are indicated.