INTRODUCTION
The adoption of molecular techniques in taxonomy has revealed unexpected genetic diversity within species throughout the tree of life (Bickford et al. Reference Bickford, Lohman, Sohdi, Ng, Meier, Winker, Ingram and Das2007). This has been especially true for the Digenea, a large group of internal metazoan parasites of animals characterized by a substantial interspecific homogeneity of the morphological characters used for species discrimination. Although the discovery of cryptic species through molecular means does not require a priori knowledge of the morphological variation in the system under study (e.g. Donald et al. Reference Donald, Kennedy, Poulin and Spencer2004; Miura et al. Reference Miura, Kuris, Torchin, Hechinger, Dunham and Chiba2005; Leung et al. Reference Leung, Keeney and Poulin2009), such knowledge is important for revealing characters that aid future identifications and thus promote a taxonomic change. Moreover, whereas many studies report the occurrence of unique genetic strains, comparatively few include formal taxonomic revisions, as such changes require a significant understanding of the morphology and systematic history of the group in question. In the taxonomy of the digeneans, as elsewhere, there has been a clear trend toward combining molecular and morphological approaches (see Nolan and Cribb, Reference Nolan and Cribb2005; Olson and Tkach, Reference Olson and Tkach2005) and this, along with increased sampling effort in recent years, has resulted in the discovery of cryptic species and higher taxa in at least 19 digenean families (e.g. Jousson and Bartoli, Reference Jousson and Bartoli2001; Nolan and Cribb, Reference Nolan and Cribb2004, Reference Nolan and Cribb2006; Chambers and Cribb, Reference Chambers and Cribb2006; Miller and Cribb, Reference Miller and Cribb2007a, Reference Miller and Cribbb; see Nolan and Cribb, Reference Nolan and Cribb2005 for a recent review).
The Haploporinae Nicoll, 1914 (Digenea: Haploporidae) is a small group of poorly known haploporid digeneans which parasitize marine or brackishwater mugilid fishes (Mugilidae). Most species were described from the Mediterranean by Looss (Reference Looss1902) and this resulted in the erection of the bulk of the haploporine genera from this area. The most ‘species-rich’ of these genera, SaccocoeliumLooss, Reference Looss1902, was erected for 2 species, S. obesumLooss, Reference Looss1902 (type-species) and S. tensumLooss, Reference Looss1902, discovered in the Adriatic Sea. Further studies have described and assigned 8 species to this genus: 6 nominal plus 2 transferred from LecithobotrysLooss, Reference Looss1902 (see Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e for details). However, most Saccocoelium spp. are only known from their original descriptions and, although the historically ‘oldest’ species described by Looss are the most widely reported in virtually all Mediterranean mullets, there are few records providing data on their morphology, and many authors have considered them synonymous (Dawes, Reference Dawes1947; Mikailov, Reference Mikailov1958; Fischthal and Kuntz, Reference Fischthal and Kuntz1963; Ferreti and Paggi, Reference Ferretti and Paggi1965; Moravec and Libosvárský, Reference Moravec and Libosvárský1975).
Until recently, no attempts have been made to address the taxonomic diversity and morphological variability within the Haploporidae and this perhaps has led to the fact that a hypothesis for a single polymorphic species has been suggested in the case of Mediterranean representatives of 3 of the 5 genera of the Haploporinae (synonymies discussed in detail by Blasco-Costa et al. Reference Blasco-Costa, Gibson, Balbuena, Raga and Kostadinova2009b, Reference Blasco-Costa, Montero, Balbuena, Raga and Kostadinovac, Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinovae). In a series of taxonomic studies based on newly-collected material from mullets in the Mediterranean basin and a critical evaluation of the published data, we have re-assessed the status of the constituent species of the ‘Mediterranean’ genera of the Haploporinae erected by Looss (Reference Looss1902) ; these studies have resulted in the erection of 3 new genera and descriptions of 4 new species (Blasco-Costa et al. Reference Blasco-Costa, Gibson, Balbuena, Raga and Kostadinova2009b, Reference Blasco-Costa, Montero, Balbuena, Raga and Kostadinovac, Reference Blasco-Costa, Montero, Gibson, Balbuena and Kostadinovad). The revision of Saccocoelium, in particular, led to the descriptions of 2 new species from Mugil cephalus L. (Saccocoelium cephali Blasco-Costa et al., Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009 and S. currani Blasco-Costa et al., Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009), re-descriptions of S. obesum and S. tensum based on material from Liza spp. and the discrimination of the 4 species from the Mediterranean with the aid of multivariate analyses. We considered S. portsaidensis El-Shahawi, El-Gindy, Imam & Al-Bassel, 1992, S. saoudi El-Shahawi, El-Gindy, Imam & Al-Bassel, 1992 and Neosaccocoelium aegyptiacus El-Shahawi, El-Gindy, Imam & Al-Bassel, 1992 to be synonyms of S. tensum, thus indicating a wide intraspecific morphological variation within this species (Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e). Furthermore, in our samples from the western Mediterranean, we could distinguish morphologically 4 morphotypes of the latter species, 2 morphotypes of S. obesum and a third form, Saccocoelium sp., parasitizing M. cephalus, which, although not identified to the species level, differed morphologically from the species of Saccocoelium that we considered valid. These findings, in combination with the unexpected diversity of haploporids revealed in M. cephalus (see also Blasco-Costa et al. Reference Blasco-Costa, Montero, Balbuena, Raga and Kostadinova2009c) prompted us to corroborate our findings using molecular data. Here we use morphological and DNA sequence data of the 28S and ITS2 rRNA genes to elucidate the taxonomic status of Saccocoelium spp. parasitizing mullets in the western Mediterranean. In particular, we test for the presence of cryptic species via the assessment of a series of isolates of the suspected morphotypes of S. obesum, S. tensum and S. cephali and examine the phylogenetic affinities of the forms parasitizing M. cephalus and Liza spp. The system selected for study is small, but characteristic for the known size of the genera of the Haploporidae, and thus provides an example for the application of a combined approach to species diversity within this problematic digenean group.
MATERIALS AND METHODS
Morphological data
Four sympatric mullet species, Mugil cephalus, Liza aurata (Risso), L. ramado (Risso) and L. saliens (Risso), were collected at 3 localities along the Mediterranean coast of Spain: Ebro Delta (40°30′–40°50′N, 0°30′–1°10′E); off Santa Pola (38°00′–38°20′N, 0°10′–0°40′E); and in a brackishwater lagoon near Santa Pola. Trematodes were killed in near-boiling saline solution, fixed in 70% alcohol, stained with iron acetocarmine, dehydrated through a graded alcohol series, cleared in dimethyl phthalate and examined as permanent mounts in Canada balsam. Eight morphotypes of Saccocoelium were distinguished on morphological grounds and sequenced in the course of the study: 2 of S. obesum ex Liza spp.; 4 of S. tensum ex Liza spp.; and 2 ex M. cephalus (S. cephali and Saccocoelium sp.). The type- and voucher material is deposited in the British Museum (Natural History) Collection at the Natural History Museum, London (BMNH) (see Table 1 for Accession numbers). All measurements are in micrometres.
Table 1. Saccocoelium spp. and ougroup taxa sequenced, their hosts, localities, number of sequenced isolates for the two rDNA regions and EMBL/GenBank and BMNH Accession numbers for sequences and morphological vouchers, respectively

Statistical analyses
Multivariate statistical analyses were performed on 13 metrical variables. First, a principal components analysis (PCA) was applied to reveal the multivariate relationship between the specimens (excluding ‘morphotype 4’ of S. tensum represented by a single specimen). Secondly, a linear discriminant analysis (LDA; backward stepwise procedure) was applied to 51 specimens assigned to 7 a priori groups in order to evaluate the morphometric differences between them and to identify the variables yielding optimal separation. The squared Mahalanobis distances between the group centroids obtained in the LDA were then used to perform a test for association between genetic and morphological distances separating recognized morphotypes by applying a method based on the permutation of distance matrices. A Mantel test for matrix correlation was carried out by regressing the Mahalanobis distances in the distance matrix from the LDA on the distances in the percentage of sequence difference matrix (ITS2 region only). The significance of the best regression model was tested with a randomization approach (9999 random permutations of the dependent variable matrix) (Manly, Reference Manly1997) using RT 2.1 program (Western EcoSystems Technology, Inc., Cheyenne, Wyoming).
Molecular data
Specimens were fixed live in 100% ethanol and stored at −20°C. Specimens were subsequently transferred into 300 μl of TNES urea (10 mm Tris-HCl (pH 8), 125 mm NaCl, 10 mm EDTA, 0·5% SDS, 4 m urea). Genomic DNA (gDNA) was extracted from single specimens using a phenol-chloroform protocol as described by Holzer et al. (Reference Holzer, Sommerville and Wootten2004). Alternatively, 1 m Tris-EDTA (pH 8) buffer was used to replace ethanol from the tissue of some specimens and gDNA was extracted using Qiagen® DNeasy™ tissue kit following the manufacturer's protocol, except for the proteinase-K incubation period, which was extended overnight, and the gDNA was further concentrated to a volume of ~30 μl using Millipore Microcon® columns. Complete ITS2 rDNA sequences were amplified using primers 3S (5′-GTA CCG GTG GAT CAC GTG GCT AGT G-3′; Anderson and Barker, Reference Anderson and Barker1993) and ITS2.2 (5′-CCT GGT TAG TTT CTT TTC CTC CGC-3′; Anderson and Barker, Reference Anderson and Barker1993). Partial (domains D1–D3; ~1400 bps) 28S rDNA sequences were amplified using primers LSU5 (5′-TAG GTC GAC CCG CTG AAY TTA AGC A-3′; Littlewood et al. Reference Littlewood, Curini-Galletti and Herniou2000) and LSU1500R (5′-GCT ATC CTG AGG GAA ACT TCG-3′; Tkach et al. Reference Tkach, Grabda-Kazubska, Pawlowski and Swiderski1999) or (~1600 bps) U178 (5′-GCA CCC GCT GAA YTT AAG-3′; Lockyer et al. Reference Lockyer, Olson and Littlewood2003) and L1642 (5′-CCA GCG CCA TCC ATT TTC A-3′; Lockyer et al. Reference Lockyer, Olson and Littlewood2003). Polymerase chain reaction (PCR) amplifications were carried out using Ready-To-Go™ (Amersham Pharmacia Biotech) PCR beads (each containing ~1·5 units Taq DNA polymerase, 10 mm Tris-HCl (pH 9), 50 mm KCl, 1·5 mm MgCl2, 200 μm of each dNTP and stabilizers, including BSA), 20–70 ng of template DNA and 10 mm of each PCR primer. The following thermocycling profile was used for ITS2 rDNA amplification: denaturation of DNA (95°C for 3 min); 35 cycles of amplification (94°C for 50 sec, 54°C for 50 sec and 72°C for 1 min 20 sec); and 4 min extension hold at 72°C. The same profile but with annealing temperatures of 58°C and 56°C, respectively for the primer combinations LSU5-L1500R and U178-L1642 was applied for 28S rDNA amplification. PCR amplicons were either gel-excised or purified directly using Qiagen QIAquick™ PCR Purification Kit and cycle-sequenced from both strands using ABI BigDye™ Terminator v3.1 Ready Sequencing Kit, alcohol-precipitated, and run on an ABI 3730 automated sequencer. The PCR primers and internal primers 300F (5′-CAA GTA CCG TGA GGG AAA GTT G-3′), ECD2 (5′-CTT GGT CCG TGT TTC AAG ACG GG-3′) and LSU1200R (5′- GCA TAG TTC ACC ATC TTT CGG-3′) (Littlewood et al. Reference Littlewood, Curini-Galletti and Herniou2000) in the case of the 28S rDNA products, were used for cycle sequencing. Contiguous sequences were assembled and edited using either Bioedit v7.0.5. (©1997–2005, Hall, Reference Hall1999) or Sequencher™ (GeneCodes Corp., ver. 3.1.1) and submitted to GenBank (see Table 1 for Accession numbers).
Alignment and phylogenetic analysis
Newly generated ITS2 rDNA and partial 28S rDNA sequences of Saccocoelium spp. were aligned using MUSCLE (Edgar, Reference Edgar2004) together with the new sequences of the haploporines Haploporus benedeniLooss, Reference Looss1902 and Forticulcita gibsoni Blasco-Costa et al., 2009 (used as outgroup taxa) and adjustments made by eye using MacClade 4.08 (Maddison and Maddison, Reference Maddison and Maddison2005). Sequences for both gene fragments were concatenated and regions of ambiguous alignment were defined in a character exclusion set. Pairwise distances for each rDNA region were calculated from the trimmed (to match the shortest sequence) aligned sequences with the absolute pairwise character difference (gaps treated as missing data) and the percentage of pairwise character differences on a total of 451 and 1189 unambiguously aligned positions for the ITS2 and the 28S rDNA, respectively.
ITS2 and 28S rDNA data partitions were analysed individually and combined using the methods of maximum parsimony (MP) and Bayesian inference (BI). MP analyses were performed with PAUP* 4.0b10 (Swofford, Reference Swofford2002) using a heuristic search strategy with 1000 search replicates, random-addition taxa sampling, tree-bisection-reconnection branch-swapping, with all characters run unordered with equal weights and with gaps treated as missing data. Nodal support was estimated by bootstrap analysis (heuristic search strategy with 1000 pseudoreplicates and 100 random sequence addition each). BI analyses were conducted using MrBayes version 3.1.2 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003). Prior to the analyses, the nucleotide substitution model was estimated using ModelTest 3.06 (Posada and Crandall, Reference Posada and Crandall1998) independently for each data partition. The model GTR+Γ (general-time-reversible model including gamma distributed among-site rate variation) was estimated as the one fitting the data best. The analyses were run over 1 million generations with a sampling frequency of 100. Consensus trees with mean branch lengths were constructed after log-likelihood values and substitution parameters plateaued at approximately generation number 12700 and 6600 for the ITS2 and 28S rDNA regions, respectively; and at 6400 for the combined analysis. Nodal support was estimated as posterior probabilities (Huelsenbeck et al. Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001).
RESULTS
Morphological data
Two morphotypes of S. obesum ex Liza spp
A detailed description of S. obesum based on material of 2 morphotypes (labelled ‘large morph 1’ and ‘large morph 2’ from L. aurata in the Ebro Delta and the Black Sea, respectively) is provided in Blasco-Costa et al. (Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e). In the latter morphological study, a third morphotype (labelled ‘small morph’) was distinguished in L. aurata from the Ebro Delta, which indicated a possible existence of a cryptic species, although no new species was proposed. Examination and sequencing of additional material from L. saliens from the same locality confirmed the distinctness of the ‘small morph’, for which we propose the name S. brayi n. sp.
S. brayi n. sp
Syn. Saccocoelium n. sp. of Blasco-Costa et al. (Reference Blasco-Costa, Balbuena, Kostadinova and Olson2009a)
Type-host: Liza aurata (Risso), the golden grey mullet (Mugilidae).
Other host: Liza saliens (Risso).
Type-locality: Off the Mediterranean coast of Spain, Ebro Delta (40°30′–40°50′N, 0°30′–1°10′E).
Site: Pyloric caeca of intestine.
Type-material: Holotype BMNH 2008.10.7.77; paratypes BMNH 2008.10.7.78-83.
Representative DNA sequences: GenBank nos. FJ211234 (partial D1-D3 28S rDNA) and FJ211244 (ITS2 rDNA); these were referred to as Saccocoelium n. sp. by Blasco-Costa et al. (Reference Blasco-Costa, Balbuena, Kostadinova and Olson2009a) to avoid nomenclatural problems due to uncertainty concerning the first publication of the name.
Etymology: The species is named for Dr Rodney A. Bray of the Natural History Museum, London, in recognition of his contributions to platyhelminth taxonomy, systematics and evolution.
Description (Fig. 1A–C)
(Based on 10 whole-mounted adult specimens; measurements in Table 2). Body elongate-oval, plump, with bell-shaped concavity at posterior extremity, with bluntly rounded posterior extremity and maximum width at posterior level of ventral sucker; width 43–59% of body length. Pre-oral lobe strongly developed, 25–51. Tegument thick (5–8), armed with large (8–10) sharp spines reaching to posterior extremity. Eye-spot pigment abundant, dispersed on either side of anterior part of pharynx and oral sucker. Oral sucker spherical, with ventral aperture. Ventral sucker cup-shaped, similar in size to or slightly larger than oral sucker (sucker length ratio 1:0·87–1·09; width ratio 1:1·00–1·28), in second quarter of body. Forebody relatively short, 35–41% of body length. Pre-pharynx absent or short (PL/PHL=0–0·6); pharynx strongly muscular, elongate-oval, larger than oral sucker. Oesophagus similar in length to pharynx. Intestinal bifurcation at level of posterior margin of ventral sucker; caeca 2 sac-like, large, with thick lining of cells, end blindly in about middle of hindbody at 26–33% from posterior extremity.

Fig. 1. Saccocoelium brayi n. sp. ex Liza aurata and L. saliens. (A) Holotype (slightly flattened) ex L. aurata, ventral view with uterus in outline. (B) Paratype ex L. saliens, ventral view with uterus in outline. (C) Paratype ex L. aurata, detail of the terminal genitalia. Scale bars: A and B=200 μm; C=100 μm.
Table 2. Comparative morphometric data for Saccocoelium obesum and S. brayi n. sp.
(Standard deviations given for egg-size only.)

* Abbreviations: BW/BL (%), maximum body width as a percentage of body length; FO/BL (%), length of the forebody as a percentage of body length; HSL/VSL (%), hermaphroditic sac length as a percentage of ventral sucker length; TEND/BL (%), post-testicular field length as a percentage of body length; CEND/BL (%), post-caecal field length as a percentage of body length.
Testis single, median to dextral, subglobular, smooth, in last quarter of body; post-testicular field 9–23% of body length. External seminal vesicle saccular, subglobular, thin-walled (<2), smaller than internal seminal vesicle. Hermaphroditic sac massive, thick-walled (5–6), muscular, elongate-oval, reaches up to distance equal to length of ventral sucker posteriorly into hindbody, much longer than ventral sucker (HSL/VSL=189–227%), contains internal seminal vesicle, numerous small prostatic cells, metraterm and hermaphroditic duct. Hermaphroditic duct wide (c.51–89), faintly-muscular, thick-walled; walls lined with crescentic, sclerotized structures. Internal seminal vesicle lined by single layer of cells, saccular, elongate-oval, occupies up to half of hermaphroditic sac. Genital atrium with strongly developed muscular wall. Genital pore median, at posterior level of pharynx.
Ovary round to transverse-oval, median to dextral, separated (holotype) from or contiguous (paratypes) with hermaphroditic sac, posterior margin of ventral sucker and testis, overlapping latter dorsally in some specimens. Uterine seminal receptacle distinct; blind seminal receptacle absent. Mehlis' gland and Laurer's canal not observed. Uterus thin-walled, occupies almost entire hindbody; metraterm distinct, joins hermaphroditic duct close to seminal vesicle. Eggs numerous, operculate; developed miracidia with single eye-spot. Vitellarium 2 separated elongate-oval clusters of large follicles, lateral at level of testis. Excretory pore terminal; details of excretory vesicle not observed (masked by uterus).
Diagnosis
The present material exhibits some of the diagnostic characteristics of, and appears most close morphologically to, S. obesum, i.e. bell-shaped concavity at posterior extremity of body, bluntly rounded posterior extremity, long forebody, large pharynx and strongly-developed muscular walls of the genital atrium (Looss, Reference Looss1902; Fares and Maillard, Reference Fares and Maillard1974; Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e). However, S. obesum is characterized by a much larger elongate cylindrical body (BW/BL=25–34 vs 43–59% (mean 30 vs 48%)), distinctly longer pre-pharynx (185–304 vs 0–76; PL/PHL=0·8–2·0 vs 0–0·6) and a somewhat smaller sucker width ratio (mean 1:0·99 vs 1:1·17) due to the ventral sucker being wider than long in S. brayi n. sp. Although the measurements of the testis and eggs exhibit overlapping ranges, the upper limits in S. brayi are higher for the former (mean 149×123 vs 125×107 in S. obesum) and lower for the latter (mean 50×27 vs 53×29, see Table 2). Furthermore, the genital atrium appears more prominent in S. obesum, whereas it is less muscular in S. brayi (mean 82×106 vs 67×75) and a vesicular pars prostatica is apparently absent in the latter species. Finally, both multivariate approaches (PCA and LDA, see below) clearly indicate a separate allocation of the specimens of S. obesum (‘large morph 1’ from the western Mediterranean), which we consider as S. obesum (sensu stricto), and specimens of S. brayi. The above comparisons coupled with the consistent multivariate morphometric differentiation and observed genetic divergence (see below) support the distinct species status of S. brayi n. sp.
Four morphotypes of S. tensum ex Liza spp
In addition to the material from L. aurata and L. ramado identified and described as S. tensum in Blasco-Costa et al. (Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e), which we label as ‘morphotype 1’ here, we found 3 isolates in L. ramado (labelled as ‘morphotypes 2–4’) that closely resembled S. tensum (Fig. 2A–D). It is worth noting that ‘morphotype 1’ was collected in fish from the Ebro Delta, ‘morphotype 2’ was the only species of Saccocoelium found in the brackish water lagoon near Santa Pola, whereas ‘morphotypes 3 and 4’ were collected in hosts fished off Santa Pola.

Fig. 2. Saccocoelium tensum ex Liza aurata and L. ramado, ventral views with uterus in outline. (A) ‘Morphotype 1’ ex L. aurata. (B) ‘Morphotype 2’ ex L. ramado. (C) ‘Morphotype 3’ ex L. ramado. (D) ‘Morphotype 4’ ex L. ramado. Scale bars=200 μm.
The specimens tentatively identified as S. tensum generally showed a morphological homogeneity, but the 3 morphotypes ex L. ramado exhibited gradually increasing upper ranges for most morphometric features (Table 3), the larger 2 forms being distinctly larger and with measurements of the ventral sucker and the length of genital atrium, testis, vitelline and postcaecal fields varying outside the ranges for ‘morphotypes 1 and 2’. Specimens of all 3 morphotypes were distinctly more elongate than those of ‘morphotype 1’, as shown by the lower upper limits (30–39 vs 56%) and means (30–33 vs 42%) for the ratio BW/BL (see also Fig. 2A–D). These differences were reflected in the separation of the specimens in the multivariate analyses (see below). Although the morphometric differentiation in the 2-dimensional plane of the PCA was not as clear for the ‘morphotypes 1 and 2’, we distinguished the latter prior to sequencing in the following characters: (i) more elongate, narrower body; (ii) long forebody; and (iii) hermaphroditic sac shorter in relation to the size of the ventral sucker, which also appears large in relation to the body (Fig. 2A–B). As shown in Table 3, the specimens of ‘morphotype 2’ also exhibit larger means for the size of the pharynx, suckers, genital atrium and eggs, and the length of the pre-pharynx, oesophagus and internal seminal vesicle, but possess smaller gonads. The specimens of this morphotype were all gravid adults bearing 30–89 eggs (30, 36, 44, 48, 76, 68 and 89, respectively), and there was no correlation between the number of eggs and body size (P=0·119).
Table 3. Comparative morphometric data for the four morphotypes of Saccocoelium tensum ex Liza aurata and L. ramado
(Abbreviations as in Table 2.)

Two Saccocoelium morphotypes ex M. cephalus
We did not expect to find yet another form parasitizing M. cephalus in the Ebro Delta, an extensively sampled locality in which 2 new species, S. cephali and S. currani, were already described (Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e; unfortunately, our attempts to obtain sequences for the latter failed). However, we distinguished morphologically a third form in this host, Saccocoelium sp., which differed from S. cephali (actually described from the voucher specimens of the present molecular study; see Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e) in its distinctly larger and more elongate body (BW/BL=22–26% vs 26–42%; mean 24 vs 36%) with maximum width at the level of the ventral sucker (vs at the junction of the first and second body thirds), larger suckers, testes, ovary and vitelline masses, and somewhat smaller eggs (Fig. 3A–B; Table 4). The specimens of Saccocoelium sp. also possessed a longer oesophagus, more anteriorly terminating caeca (at mid-body vs mid-hindbody) and a more anterior location of the most posterior uterine loops (UEND/BL=22–42 vs 8–16%). The size differences could not be attributed to growth, since some of the specimens of the larger form, Saccocoelium sp., were neogravid. However, lack of sufficient numbers of fully-gravid worms prevented confident identification of the species.

Fig. 3. Saccocoelium morphotypes ex Mugil cephalus. (A) S. cephali, ventral view of a paratype with uterus in outline. (B) Saccocoelium sp., ventral view with uterus in outline. Scale bars=200 μm.
Table 4. Comparative morphometric data for Saccocoelium cephali and Saccocoelium sp. ex Mugil cephalus
(Abbreviations as in Table 2.)

Morphometric variation of Saccocoelium spp
The considerable morphological variability encountered in the present collection required a thorough examination to determine the species status of the morphotypes. We, therefore, applied the approach adopted in previous studies on the Haploporinae (see Blasco-Costa et al. Reference Blasco-Costa, Montero, Balbuena, Raga and Kostadinova2009c, Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinovae). First, PCA was applied to assess and visualize the overlap of the morphological characteristics of the Saccocoelium spp. The first two principal components of the PCA run on the correlation matrix between 13 metrical variables of the 8 morphotypes explained 71·6% of the variation in the data-set comprising 51 specimens. The size of the suckers and the length of the forebody had the highest coefficients on the first component, which explained 56% of the total variance (eigenvalue 7·8), whereas testis size and the length of post-uterine field had important contributions to the second principal component, which explained a further 15·6% of the variance (eigenvalue 2·2). A plot of the specimens in the first plane of the PCA (Fig. 4A) shows, along both the first and second axis, 2 well-separated groups that correspond to the 2 morphotypes from M. cephalus. Specimens of S. obesum (sensu stricto) and S. brayi n. sp. appeared close, but separated along the first axis. On the other hand, the morphotypes of S. tensum did not show a clear clustering pattern. The specimens of S. brayi n. sp. appeared closer and overlapped morphotypes of S. tensum in this analysis based on morphometric data only. However, the new species can be readily distinguished from S. tensum using only the structure of the posterior extremity of the body.

Fig. 4. Plots of the 52 specimens of Saccocoelium spp. (A) In the first plane of the PCA. (B) Against the first and second canonical discriminant functions (LDA). Key to morphotypes of S. tensum: ‘Morphotype 1’, open circles; ‘Morphotype 2’, light grey circles; ‘Morphotype 3’, dark grey circles.
Secondly, the backward stepwise LDA procedure run on 13 metrical variables separated the 7 morphotypes with 92% accuracy (Fig. 4B) (Wilk's Lambda=0·00121; approximate F(24, 144)=35·2, P<0·0001). The first canonical function clearly discriminates the two morphotypes ex M. cephalus and S. obesum (s. s.) from the remaining specimens, whereas the second canonical function contributes to the discrimination between the two morphotypes from M. cephalus, between S. obesum (s. s.) and S. brayi n. sp. and between the former and the morphotypes of S. tensum. The specimens of S. obesum (s. s.), ‘morphotype 1’ of S. tensum and the two morphotypes from M. cephalus which formed distinct clusters in Fig. 4B were all correctly assigned to their a priori groups whereas 2 specimens of S. brayi n. sp. were misclassified as ‘morphotype 1’ of S. tensum (accuracy 71%) and 1 specimen of ‘morphotype 2’ and ‘morphotype 3’ of S. tensum each were misclassified as S. brayi (accuracy of 85 and 67%, respectively); these uncertainties are indicated by the higher dispersion and overlap among the samples of S. tensum and S. brayi. The discriminatory power of the model was associated with only 4 variables. The length of the testis and the length of the muscular genital atrium exhibited strong correlation with the first axis, and the length of the forebody and the width of the ventral sucker were associated with the discrimination along the second axis of the 2-dimensional plane.
Molecular analysis
Sequences of both 28S and ITS2 rRNA gene regions were obtained from a total of 21 isolates of the 8 morphotypes of Saccocoelium spp. Sequences for the 2 rDNA regions from multiple isolates of S. tensum, S. obesum and Saccocoelium sp. (Table 1) showed no variation within the sequences for either region. The 28S and ITS2 sequences for all morphotypes examined were aligned together with the sequences for the two outgroup taxa. The alignment of the 28S rDNA incorporated a total of 1189 included characters (bp and gaps) and the alignment of the ITS2 represented a total of 451 characters. Comparative sequence analysis revealed 4 unique genotypes for each of the two rDNA regions examined: (i) S. obesum (s. s.); (ii) S. brayi n. sp.; (iii) S. tensum (all 4 morphotypes); and (iv) S. cephali/Saccocoelium sp.
Divergence in the 28S sequence between species of Saccocoelium ranged from 0·8 to 4·3% (9–51 nucleotide sites) and those in the ITS2 sequence ranged from 1·6 to 7·2% (7–33 nt) (Table 5). The smallest divergences were observed between the two morphotypes of S. obesum (sensu lato) [i.e. S. obesum (s. s.) and S. brayi n. sp.], both showing the highest percentage of sequence difference to the other two Saccocoelium genotypes. However, the two morphotypes from M. cephalus, which appeared morphologically distinct (S. cephali and Saccocoelium sp., see above), shared the same genotype for both regions analysed and the 4 morphotypes of S. tensum (1 from L. aurata and 3 from L. ramado) were genetically identical for the 2 rDNA regions. The Mantel test, aimed to assess the congruence between the genetic (ITS2 region) and morphometric differentiation between the 7 morphotypes, resulted in a high P-value (P=0·32), thus confirming the lack of correlation between the genetic and morphometric distance matrices in the present material.
Table 5. Pairwise nucleotide sequence comparisons between taxa, calculated as percentage of nucleotide differences (gaps treated as missing data) for the aligned ITS2 (above the diagonal; N=451 bps) and 28S rDNA (below the diagonal; N=1189 bps) sequences

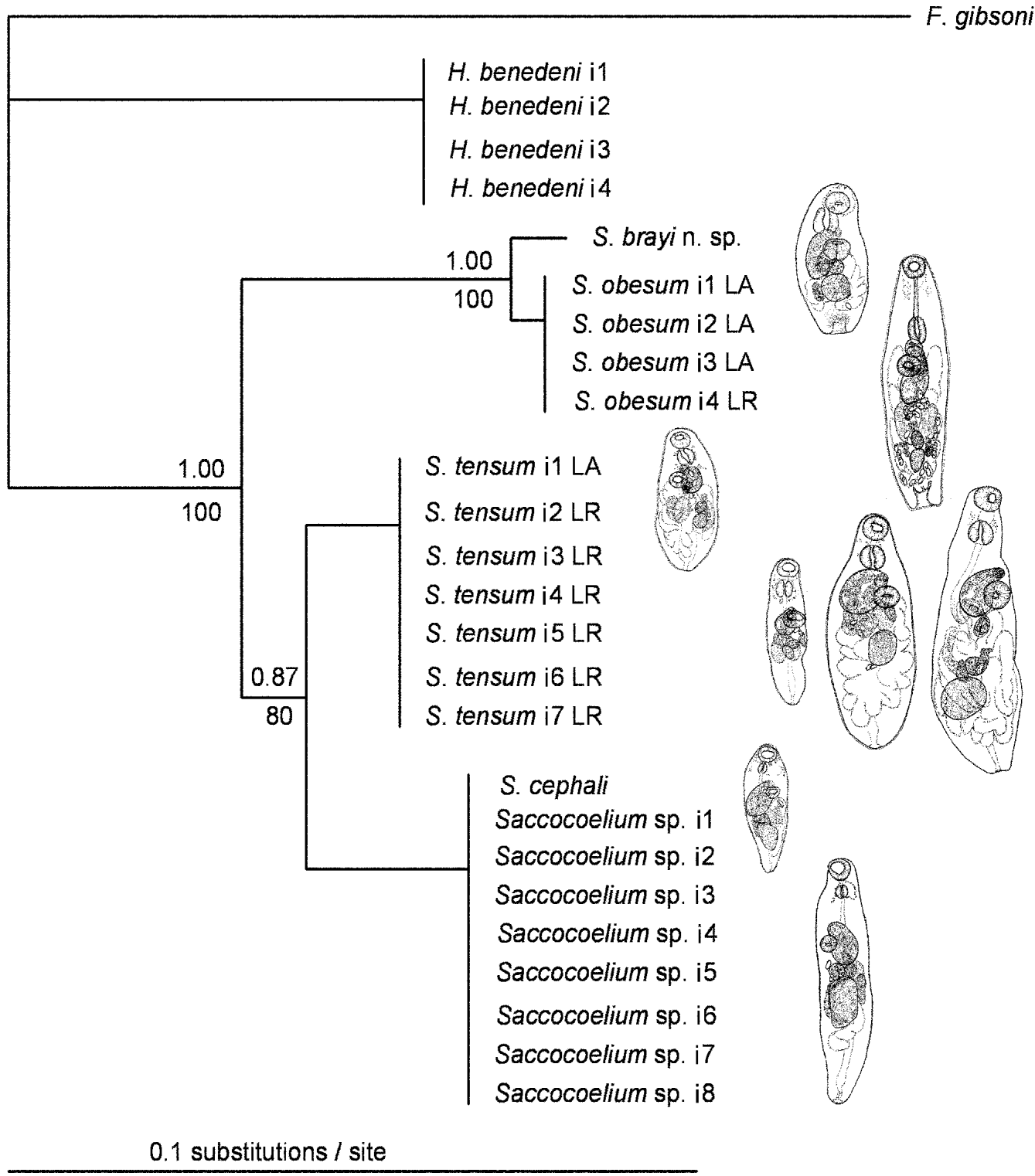
Figure 5 presents the phylogram generated with BI analyses of the combined dataset (28S and ITS2). MP of the combined dataset produced a single most-parsimonious tree (length 336, consistency index 0·896) with strong nodal support for all clades (not shown). The tree topologies, obtained in independent analyses of the 2 gene regions, shared the same branching patterns and showed similar levels of support. Within the ingroup, 2 strongly supported clades were recognized, one formed by morphotypes of S. tensum from L. aurata and L. ramado and morphotypes from M. cephalus, and the other comprising S. obesum (s. s.) and S. brayi n. sp., the latter being subtended by a longer branch.

Fig. 5. Tree topology derived from the combined 28S and ITS2 rRNA gene sequences using Bayesian analysis with posterior probability values above and MP bootstrap values below the branches. Abbreviations: i followed by number, sequence isolate number; LA, Liza aurata; LR, Liza ramado.
DISCUSSION
Our study is the first parallel molecular and morphological attempt focused on characterization of a group of congeneric species within the Haploporidae, a poorly known digenean family characterized by a long history of inadequate descriptions, poor specific diagnoses, scattered records and extensive synonymy (Overstreet and Curran, Reference Overstreet, Curran, Jones, Bray and Gibson2005). The family has also been found to be highly labile in its placement in the most comprehensive molecular phylogenetic analysis of the Digenea performed to date (Olson et al. Reference Olson, Cribb, Tkach, Bray and Littlewood2003), and this reflects the difficulty in characterizing the group at effectively all taxonomic levels. Combining sequence data analysis with a detailed morphological and multivariate morphometric study of the specimens has allowed the demonstration of cryptic diversity among the closely related species of Saccocoelium. One important result is that molecular data corroborate our decisions based on morphology (Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e) with respect to the distinct status of 3 species, S. obesum (s. s.), S. tensum and S. cephali, thus rejecting the hypothesis of a single species infecting sympatric mullets in the Mediterranean (e.g. Dawes, Reference Dawes1947; Mikailov, Reference Mikailov1958; Fischthal and Kunz, Reference Fischthal and Kuntz1963; Ferreti and Paggi, Reference Ferretti and Paggi1965; Moravec and Libosvárský, Reference Moravec and Libosvárský1975). Furthermore, the analysis of both gene regions supported the recognition of another cryptic species, S. brayi n. sp. Species recognition based on sequence divergence in the ITS2 in cases when low interspecific variation is detected between congeners is difficult; one is doomed to either fail to recognize multiple cryptic species or provide ‘species’ destined for synonymy (see Nolan and Cribb, Reference Nolan and Cribb2005 for a review). The amount of morphological and genetic variation between S. obesum (s. s.) and S. brayi n. sp. was lower than that observed between S. tensum and S. cephali, and this would suggest their recent separation. In addition, the morphological differences between the morphotypes of S. obesum (s. l.) were confirmed by the multivariate morphometric analyses. These, coupled with the observed sequence divergence in the 28S and ITS2 regions, the former slightly above the minima (e.g. 0·2–0·4% in the Cryptogonimidae, see Miller and Cribb, Reference Miller and Cribb2007a, Reference Miller and Cribbb) and the latter higher than or closely approaching the lowest levels reported between congeneric taxa in other marine digenean systems (e.g. 0·5% in the Didymozoidae, see Anderson and Barker, Reference Anderson and Barker1998; 0·3% in the Sanguinicolidae, see Nolan and Cribb, Reference Nolan and Cribb2006; 0·4–1·4% in the Cryptogonimidae, see Miller and Cribb, Reference Miller and Cribb2007a, Reference Miller and Cribbb), support our decision to recognize the distinct species status of S. brayi n. sp. Further sequencing, especially of material from the Black Sea, will be useful to test our hypothesis for higher diversity in the S. obesum (s. l.) complex (Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e) and to reveal the divergence rates within this morphologically and genetically distinct lineage of Saccocoelium.
The lack of genetic differentiation among the 4 morphotypes of S. tensum is in accord with our initial hypothesis of a single morphologically variable species based on the long list of synonyms and intraspecific variation detected in descriptions from various mullet hosts (Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e), and the lack of qualitative differentiating features and the gradually overlapping ranges for the metrical data in the present material. However, the finding that Saccocoelium sp. isolates had identical sequences to S. cephali, especially of the more variable ITS2 region, was unexpected in view of the apparent boundaries indicated by comparative morphology and multivariate statistical analysis; this is reflected in the larger number of isolates sequenced. One explanation for the observed lack of genetic differentiation in the ITS2 region within the S. tensum – S. cephali clade may be a slower rate of evolution than that found in the S. obesum (s. s.)+S. brayi n. sp. lineage, the latter exhibiting a considerably longer branch length. If this is the case, it is possible that the ITS2 region may not provide enough resolution to guarantee distinguishing between all combinations of closely related haploporid species (see also Nolan and Cribb, Reference Nolan and Cribb2005). Nevertheless, ensuing from the fact that to date just a single convincing example for identical ITS2 sequences in genuinely different species exists within the Digenea (Blair et al. Reference Blair, Agatsuma, Watanobe, Okamoto and Ito1997; discussed in Nolan and Cribb, Reference Nolan and Cribb2005), we adopted a conservative approach, considering the distinct species status only for the Saccocoelium isolates that are supported by both morphological and molecular evidence. However, we provide sufficient data for their morphological differentiation and have deposited voucher material of the morphotypes studied here, in order to enable their recognition should future studies on different loci (e.g. ITS1 or mitochondrial genes) offer evidence validating their species distinction.
Divergence of the two clades of Saccocoelium appears to be associated with a change in the intermediate host group rather than with diversification related to the definitive hosts. Life-cycle data available for S. tensum and S. obesum (if extended to the other members of their respective clades) tend to support this suggestion. Cercariae of S. tensum develop in the hydrobiid snails Hydrobia acuta and H. ventrosa, whereas those of S. obesum develop in the rissoid snails Rissoa spp. (Fares and Maillard, Reference Fares and Maillard1974; Deblock, Reference Deblock1980; the former authors also reported numerous unsuccessful attempts to infect experimentally Hydrobia spp. with S. obesum). The free-living cercariae of both species encyst in the open by attaching to the substrate and are thus available for passive ingestion by the definitive pump-filtering, detritivorous (Cardona et al. Reference Cardona, Royo and Torras2001) mullet hosts in which the adult stages develop. Host-parasite data, although recently updated, are still wanting due to the large body of non-documented records (i.e. with no supportive evidence for the species identification). Nevertheless, documented records for both S. obesum and S. tensum include 5 of the 6 widespread sympatric Mediterranean mullet species (i.e. Chelon labrosus, Mugil cephalus, Liza aurata, L. ramado and L. saliens) as definitive hosts (see Blasco-Costa et al. Reference Blasco-Costa, Montero, Gibson, Balbuena, Raga and Kostadinova2009e for details). This fact and the current lineage division suggest that parasite specificity at the level of the definitive host may not serve as an important force for speciation in the mullet-haploporid system studied. This situation is similar to that observed in a Mediterranean sparid-digenean system comprising 3 host species which occur in sympatry (Jousson et al. Reference Jousson, Bartoli and Pawlowski2000). A wide open encounter filter (sensuCombes, Reference Combes1995; Combes and Théron, Reference Combes and Théron2000, Poulin, Reference Poulin2006) in our system resulting from the combination of the distinctly ‘passive’ transmission of the infective metacercariae which are ingested non-selectively and the habitat/trophic overlap between the mullet host species would tend to reduce the possibility of co-existence of genetically isolated mullet-specific lineages of Saccocoelium.
However, the higher than previously thought species diversity, confirmed by the present molecular data at the limited geographical scale of the study (c. 500 km, with parasite species co-occurring in the same locality) and the coexistence of divergent parasite lineages in the same host species indicate the action of factors linked to the haploporid life-cycle which modify the effect of the enhanced encounter with the definitive hosts. Two non-exclusive hypotheses can be suggested for the observed patterns of species and genetic diversity in our system. The first, and an obvious prediction, is that the existence of more species of Saccocoelium reflects adaptation to different sympatric snail hosts. The relationship with the snail intermediate hosts can be viewed as the most important component of the compatibility filter (sensuCombes, Reference Combes1995, Poulin, Reference Poulin2006), since digeneans exhibit the highest specificity to their molluscan hosts (Pearson, Reference Pearson1972; Adamson and Caira, Reference Adamson and Caira1994), and this supports our suggestion. Estimation of both hydrobiid and rissoid species diversity in the Mediterranean is difficult due to problems in species delimitation (Anistratenko and Stadnichenko, Reference Anistratenko and Stadnichenko1994; Wilke and Davis, Reference Wilke and Davis2000 and references therein). However, a particularly rich set of sibling species was established in European rissoaceans (Russo and Patti, Reference Russo and Patti2005 and references therein) and members of Hydrobia were shown to represent morphostatic species radiations (i.e. ‘… considerable, rapid speciation[s] with low anatomical diversification’; see Davis, Reference Davis1994; Wilke and Davis, Reference Wilke and Davis2000). It is therefore possible that speciation within Saccocoelium is associated with cryptic diversification of the first intermediate rissoid and hydrobiid hosts. Sequencing of both snail hosts and larval stages in the western Mediterranean lagoons would provide a test of this hypothesis.
Our second hypothesis is that the higher parasite diversity is a result of local adaptation governed by larval dispersal of the snail intermediate hosts. Larval spatial distributions and dispersal ability have been linked to genetic differentiation among free-living marine organisms (e.g. Tatarenkov and Johannesson, Reference Tatarenkov and Johannesson1998; Boisselier-Dubayle and Gofas, Reference Boisselier-Dubayle and Gofas1999; Riginos and Victor, Reference Riginos and Victor2001). Of particular relevance to the system under study is the fact that H. ventrosa (first intermediate host of S. tensum and 2 other haploporid species) is a species with a direct development (i.e. crawl-away juveniles emerge after metamorphosis from egg masses deposited on the substrate), and this results in high population level differentiation and low gene flow between populations (Foltz, Reference Foltz2003; Wilke and Davis, Reference Wilke and Davis2000). The poor dispersal and the heterogeneity of the habitats (see Bartoli and Gibson, Reference Bartoli and Gibson2007 for comment on lagoonal types in the western Mediterranean) may therefore provide a setting for the development of differential susceptibility in the populations of this host towards infections with Saccocoelium spp. and haploporids in general. Thus, the possibility of speciation via local adaptation, resulting from the spatial structure of the first intermediate host populations, might be high in haploporids.
Summarizing the results of our study, we conclude that distinct species status is only valid for the Saccocoelium isolates that are supported by both morphological and molecular evidence. Our data suggest that delimiting species using solely morphological criteria may be misleading; however, we do not rule out the possibility for even higher species diversity within the studied digenean group. By describing sequence and morphological divergence across the lowest taxonomic levels, we provide a test case that demonstrates which genetic and morphological markers can be used for diagnostic analysis in the Haploporidae.
We are grateful to David Gibson, Natural History Museum, London, for advice in the course of the study and to Tomas Scholz, Institute of Parasitology, Biology Centre of the Academy of Sciences of the Czech Republic, for comments on the manuscript. This research was supported by INTAS (J.A.B., A.K., grant 03-51-5998); Ministry of Science and Education of Spain (J.A.B., grant CGL2008-02701); FP6-MTKD (J.A.R., A.K., grant CT-2004-003175); and Institute of Parasitology (A.K., grants Z60220518 and LC522). I.B.-C. benefits from a doctoral fellowship from the Ministry of Science and Education of Spain (AP-2004-2999) and a visiting studentship to the Natural History Museum in London.