Introduction
More than 50 species have been found to belong to the genus Bacidina since it was first described by Vězda (Reference Vězda1990). They were transferred to this genus (e.g. Vězda Reference Vězda1990; Wirth Reference Wirth1994; Ekman 1996 Reference Ekmana , Reference Ekman2004; Hauck & Wirth Reference Hauck and Wirth2010; Farkas Reference Farkas2015) or were newly described (e.g. Farkas & Vězda Reference Farkas and Vězda1993; Ekman 1996 Reference Ekmana ; Cáceres Reference Cáceres2007; Spribille et al. Reference Spribille, Björk, Ekman, Elix, Goward, Printzen, Tønsberg and Wheeler2009; van den Boom & Sipman Reference van den Boom and Sipman2014) mainly based on morphological and anatomical characters, more particularly biatorine apothecia; excipular radiating hyphae with at least apically large, broadly ellipsoid to almost globose lumina (±paraplectenchymatous excipulum at least in an external portion); narrowly clavate asci with a distinct axial body that is basically conical but varies in height and width; and filiform, long, septate, usually curved conidia (for more details see Ekman 1996 Reference Ekmana ).
To date only a small proportion of validly described Bacidina species (10 species) have been included in phylogenetic analyses (Ekman Reference Ekman2001; Czarnota & Guzow-Krzemińska Reference Czarnota and Guzow-Krzemińska2012) and consequently molecular affinities within the genus remain unresolved. Further approaches are needed to address this due to morphological variation which obscures the boundaries of many species. Such variability is found especially in representatives of Bacidina which are often labelled as Bacidina neosquamulosa (Aptroot & Herk) S. Ekman (or Bacidia neosquamulosa Aptroot & Herk), Bacidina adastra (Sparrius & Aptroot) M. Hauck & V. Wirth (or Bacidia adastra Sparrius & Aptroot) and Bacidina caligans (Nyl.) Llop & Hladún (or Bacidia caligans (Nyl.) A. L. Sm.). It was clear that the application of molecular data is necessary to understand relationships within this group of species and support potential nomenclatural innovations.
The main objectives of this study were to clarify species boundaries and relationships among specimens previously identified as Bacidina neosquamulosa or B. caligans, and to improve our understanding of the phylogenetic relationships within the genus Bacidina.
Materials and Methods
Morphology and chemistry
All specimens examined were studied with a Zeiss Stemi DV4 stereomicroscope and a Zeiss Axiostar plus light microscope. Hand-cut apothecial sections mounted in water were prepared for measuring anatomical characters and photographs; at least 20 measurements were made for each diagnostic feature. We used 10% KOH to separate apothecial structures and to examine internal pigment reactions, and Lugol’s iodine for the examination of ascus structure. The presence of secondary metabolites was studied using a standard TLC method with solvent C (Orange at al. Reference Orange, James and White2001).
Taxon sampling for DNA analyses
Specimens of Bacidina used for DNA analysis are listed in Table 1 and their GenBank Accession numbers are provided; these and other sequences downloaded from GenBank were used in the phylogenetic analyses. In total, 24 new sequences of ITS rDNA were obtained, including that for the newly described B. mendax. Usnea fragilescens and U. subantarctica were used as an outgroup in the phylogenetic analyses.
Table 1 Voucher information and GenBank Accession numbers for the new ITS rDNA sequences used in the phylogenetic analysis, including the holotype of Bacidina mendax sp. nov. named as B. neosquamulosa in GenBank
DNA extraction, PCR amplification and sequencing of the ITS rDNA region
DNA was extracted directly from pieces of thallus using a modified CTAB method (Guzow-Krzemińska & Węgrzyn Reference Guzow-Krzemińska and Węgrzyn2000). DNA extracts were used for PCR amplification of the ITS rDNA marker using the primers ITS1F (Gardes & Bruns Reference Gardes and Bruns1993) and ITS4 (White et al. Reference White, Bruns, Lee and Taylor1990). The 25 μl of PCR mix contained 1U of Taq polymerase (Thermo Scientific), 0·2 mM of each of the four dNTPs, 0·5 μM of each primer and 10–50 ng of genomic DNA. PCR amplifications were performed using a Mastercycler (Eppendorf) with the following programme: initial denaturation at 95°C for 5 min followed by 35 cycles at 95°C for 40 s, 54°C for 45 s and 72°C for 1 min, and a final elongation step at 72°C for 10 min. PCR products were visualized on agarose gels in order to determine DNA fragment lengths. Subsequently, 5 µl of PCR product was treated with 10 units of Exonuclease I and 1 unit of FastAPTM Thermosensitive Alkaline Phosphatase enzymes (Thermo Scientific) to degrade primers and dephosphorylate dNTPs. Treatment was carried out for 15 min at 37°C, followed by a 15 min incubation at 85°C to completely inactivate both enzymes. Sequencing of each PCR product was performed using Macrogen sequencing service (www.macrogen.com).
Sequence alignment and phylogenetic analysis
The newly generated ITS rDNA sequences were compared with those available in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) using a BLASTN search (Altschul et al. Reference Altschul, Gish, Miller, Myers and Lipman1990) in order to confirm their identity. The ITS rDNA sequences were aligned with sequences of selected representatives of the genera Bacidina and Bacidia, as well as Biatora, Bilimbia, Toninia and Usnea (GenBank Accession numbers provided in Fig. 1). Alignment was performed using MAFFT with the parameters set to default values as available from EBI (http://www.ebi.ac.uk/Tools/msa/mafft/) (Katoh et al. Reference Katoh, Misawa, Kuma and Miyata2002) and followed with a selection of ambiguous positions that might not have been homologous using Gblocks 0.91b, implementing the options for less stringent parameters (i.e. to allow less strict flanking positions and smaller final blocks) (Castresana Reference Castresana2000; Dereeper et al. Reference Dereeper, Guignon, Blanc, Audic, Buffet, Chevenet, Dufayard, Guindon, Lefort and Lescot2008).
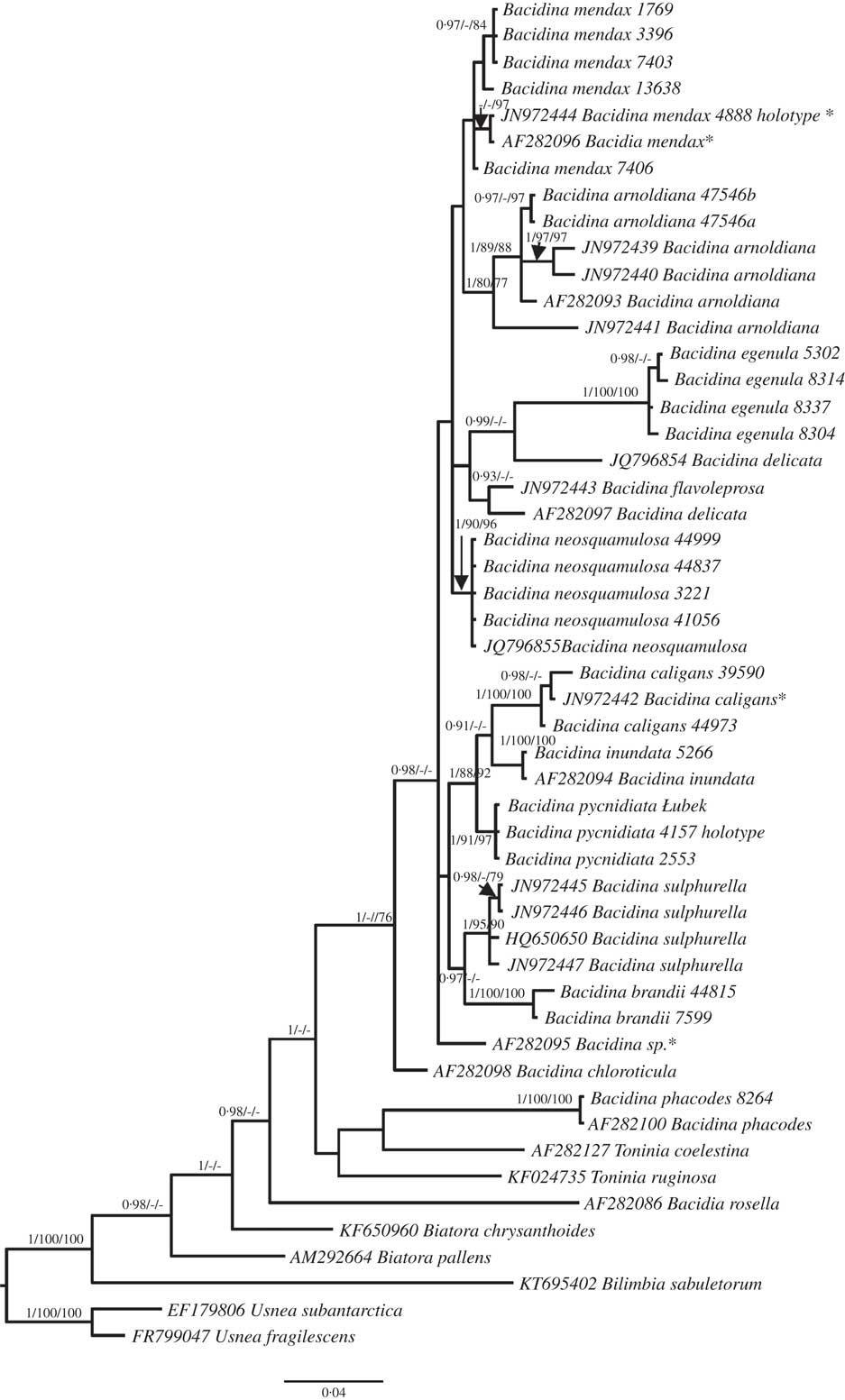
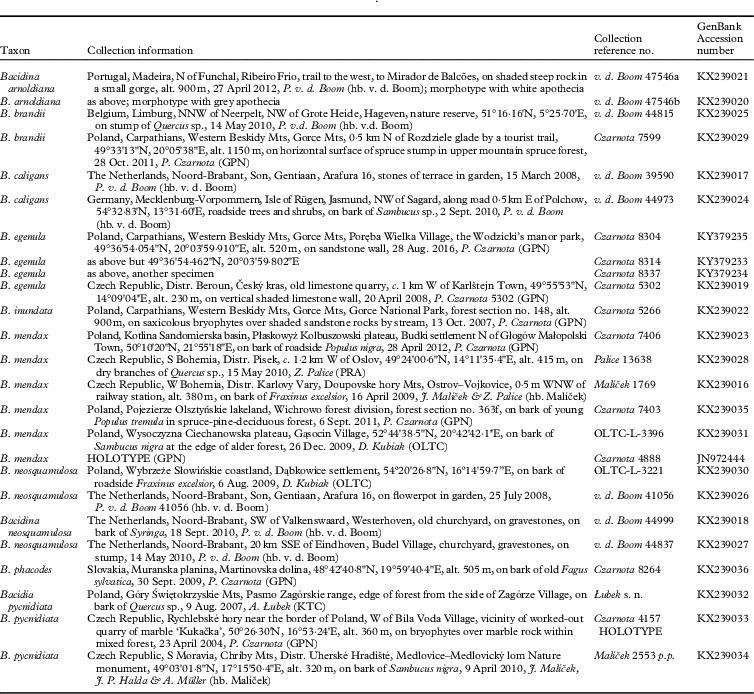
Fig. 1 Bayesian tree based on ITS rDNA data for Bacidina spp. Usnea fragilescens and U. subantarctica are the outgroup taxa. Support values are displayed near the branches in the following format: posterior probabilities >0·90 /bootstrap percentage for MP (maximum parsimony) >75 / ML (maximum liklehood) >75. Species names are followed by their GenBank Accession numbers, apart from newly sequenced specimens of Bacidina spp. where the names are followed by herbarium collection numbers. *=taxa that were labelled with other names in GenBank (i.e. JN972444 as B. neosquamulosa, AF282096 as B. caligans, AF282095 as B. egenula and JN972442 as B. adastra).
The phylogenetic analyses were performed using PAUP* 4.0b10 (Swofford Reference Swofford2001) with maximum parsimony (MP) as the optimality criterion. Heuristic searches were performed with 1000 random sequence additions and TBR branch swapping. Gaps were treated as missing and support for the branches was evaluated with a bootstrap method with 100 pseudoreplicates (Felsenstein Reference Felsenstein1985).
Maximum likelihood (ML) analyses were performed using RaxML HPC v.8 on XSEDE (Stamatakis Reference Stamatakis2014) under the GTRGAMMAI model on the CIPRES Science Gateway (Miller et al. Reference Miller, Pfeiffer and Schwartz2010). Rapid bootstrap analyses were performed with 1000 bootstrap replicates.
The data were also analyzed using a Bayesian approach (MCMC) in MrBayes 3.2.6 (Huelsenbeck & Ronquist Reference Huelsenbeck and Ronquist2001; Ronquist & Huelsenbeck Reference Ronquist and Huelsenbeck2003). The GTR+G+I model was selected based on analysis using MrModeltest 2.0 (Nylander Reference Nylander2004). A run with 5 000 000 generations employing four chains was selected and every 100th tree was saved. The initial 25% of trees were discarded as burn-in and a majority-rule consensus tree was calculated to obtain posterior probabilities (BA).
The phylogenetic tree was drawn using FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Bootstrap supports (in MP and ML) above 75% and posterior probabilities above 0·90 (in BA) were indicated near the branches as these values were considered to be significant.
Results
Twenty-four new ITS rDNA sequences were obtained from eight representatives from the genus Bacidina (including the newly described B. mendax Czarnota & Guz.-Krzem. sp. nov.) and from Bacidia pycnidiata Czarnota & Coppins. Among them, Bacidina brandii (Coppins & van den Boom) M. Hauck & V. Wirth, B. egenula (Nyl.) Vězda and Bacidia pycnidiata were sequenced for the first time (see Table 1 for the list of all newly generated sequences).
In the dataset, 51 ITS rDNA sequences were analyzed; of 417 characters, 233 were constant and 128 were parsimony-informative. 113 equally parsimonious trees of 491 steps were generated (MP) and the majority-rule consensus tree was created (data not shown). Trees of similar topologies were also generated using the maximum likelihood (ML; best tree likelihood LnL=−2895·180637) method and Bayesian approach (BA; harmonic mean was −3160·33). In the MrBayes analysis, the average standard deviation of split frequencies was 0·004069 and the average PSRF for parameter values was 1·00. The Bayesian tree is presented in Fig. 1 with added bootstrap supports from the MP and ML methods and posterior probabilities from the BA.
Species belonging to Bacidina, including B. mendax sp. nov., form a monophyletic clade supported by bootstrap value (76 in ML) and posterior probability in BA (1·00) (Fig. 1). However, the position of B. phacodes (Körb.) Vězda, the type species of Bacidina, within the genus is not supported in all trees, and thus remains uncertain.
The morphologically variable specimens related to B. mendax (Figs 2 & 3) are grouped together, but without support (Fig. 1); they represent four different ITS haplotypes.
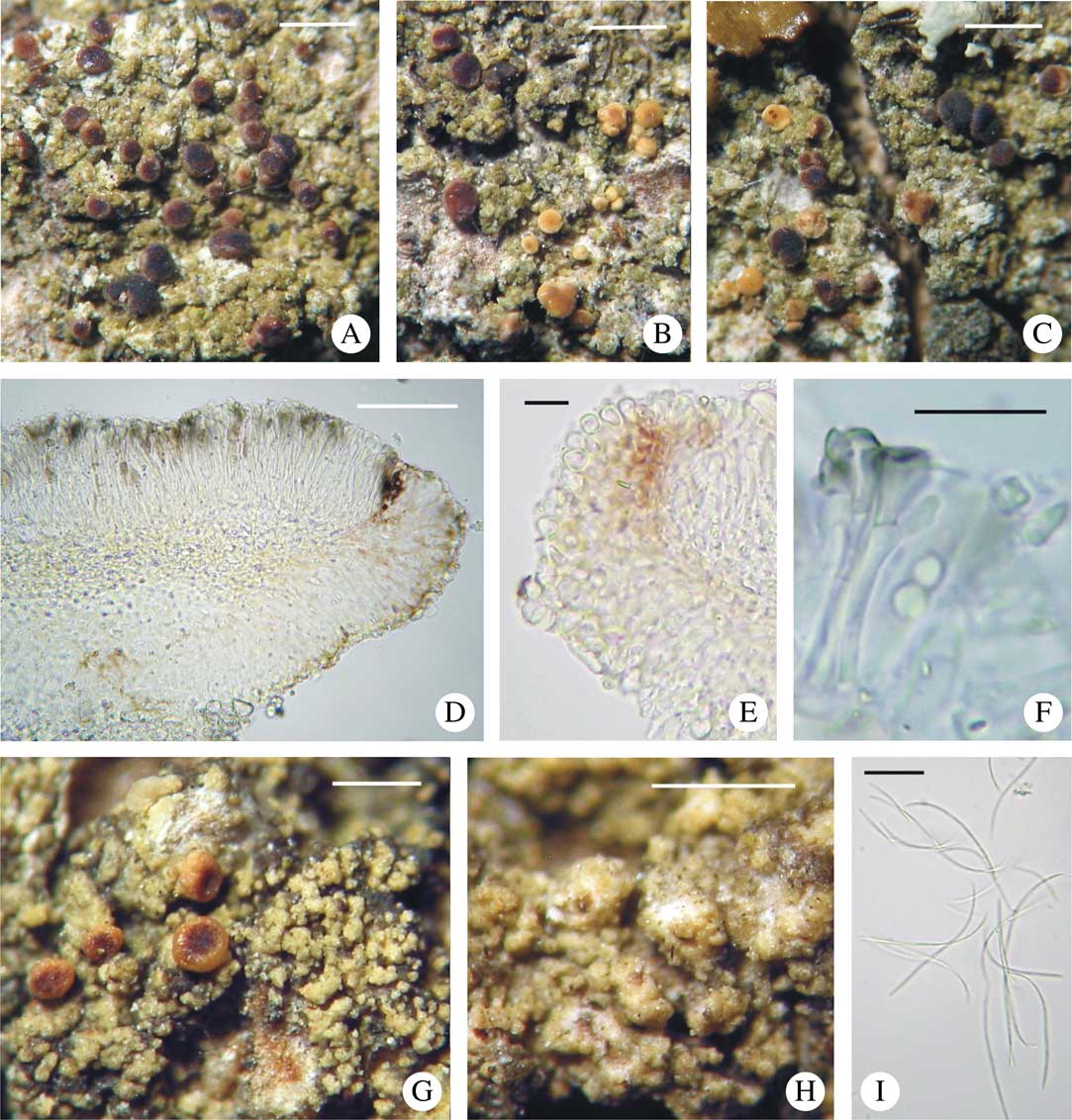
Fig. 2 Bacidina mendax. A–C & G, habitus of variously coloured morphotypes; D, section though apothecium; E, section through excipulum with external rows of globose, apical cells; F, paraphyses of the second type forming pigmented, vertical streaks (see species description); H, pycnidia; I, pycnospores. A–D & F, Czarnota 4888 (GPN—holotype, GenBank no. JN972444); E & G–I, Czarnota 7406 (GPN—paratype, GenBank no. KX239023). Scales: A–C, G & H=1 mm; D=50 μm; E & F=10 μm; I=20 μm. In colour online.
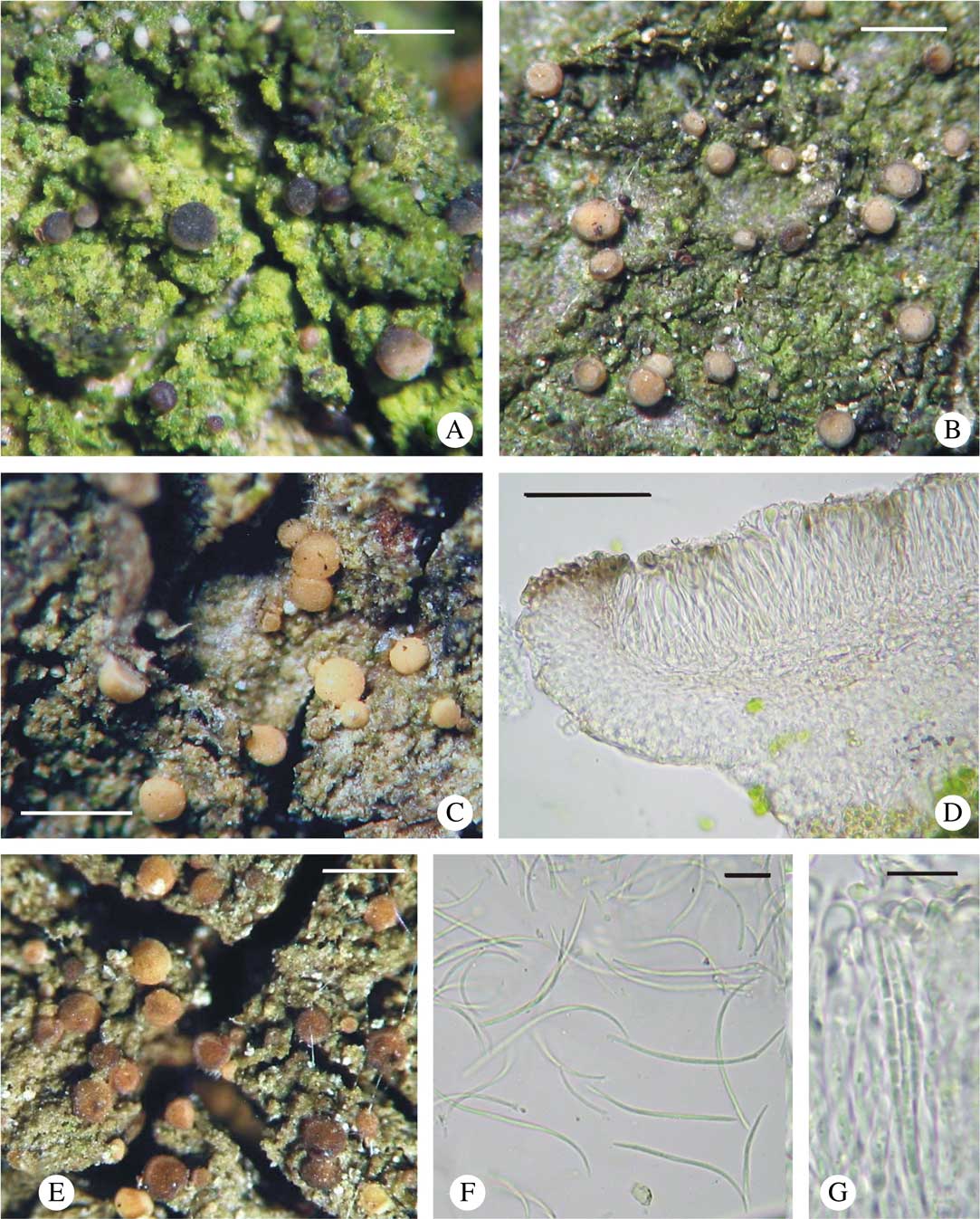
Fig. 3 Bacidina mendax. A–C & E, habitus of different morphotypes; D, section through apothecium; F, pycnospores; G, ascus with ascospores. A & D, Kubiak (OLTC-L-3396—paratype, GenBank no. KX239031); B, Łubek s. n. (KTC, loc. P1—paratype), C, Malíček 1769 (hb. Malíček—paratype, GenBank no. KX239016); E, Palice 13638 (PRA—paratype, GenBank no. KX239028); F, Czarnota 7403 (GPN—paratype, GenBank no. KX239035); G, Łubek s. n. (KTC, loc. M11—paratype). Scales: A–C & E=1 mm; D=50 μm; F & G=10 μm. In colour online.
The four newly sequenced specimens of B. egenula form a highly supported lineage (100 in MP, 100 in ML and 1·00 in BA), but the sequence no. AF282095 from GenBank named B. egenula has no relationship to the clade mentioned above (Fig. 1). However, since it is nested within Bacidina, a provisional name Bacidina sp. has been adopted.
Six specimens of B. arnoldiana (Körb.) V. Wirth & Vězda form a moderately supported clade (80 in MP, 77 in ML and 1·00 in BA) (Fig. 1), as previously shown by Czarnota & Guzow-Krzemińska (Reference Czarnota and Guzow-Krzemińska2012); ITS rDNA data show that it is a highly variable species.
Three sequences from the specimens of B. caligans form a single highly supported clade (100 in both MP and ML methods and 1·00 in BA). Their haplotypes differ from each other in one to four positions. This species was found to be related to B. inundata (Fr.) Vězda (0·91 in BA) (Fig. 1).
Newly sequenced Bacidia pycnidiata, represented here by three specimens collected in the Czech Republic and Poland, is placed within the genus Bacidina Vězda; we therefore propose to transfer Bacidia pycnidiata to Bacidina. This species was found to be related to B. inundata and B. caligans, and their relationship is supported in all optimization methods (88 in MP, 92 in ML and 1·00 in BA) (Fig. 1).
ITS rDNA sequences obtained from B. neosquamulosa represent two haplotypes that differ in a single position and form a monophyletic, highly supported clade (90 in MP, 96 in ML and 1·00 in BA) (Fig. 1); specimens are very variable in the morphology of apothecia and their thallus structure (Fig. 4).
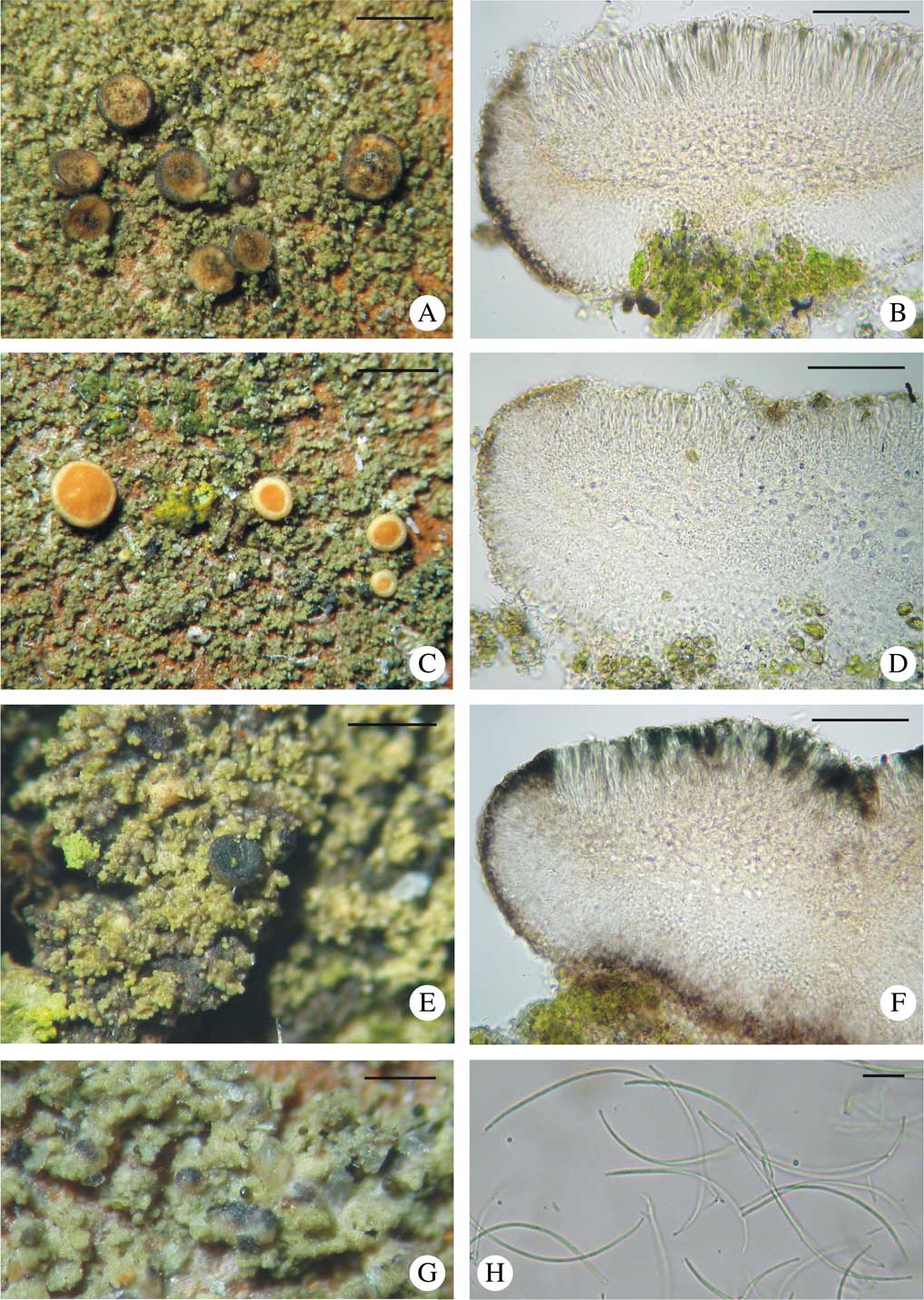
Fig. 4 Bacidina neosquamulosa. A, C & E, habitus of variously coloured morphotypes; B, D & F, sections through variously pigmented apothecia; G, pycnidia; H, pycnospores. A–D & G, v. d. Boom 41056 (hb. v. d. Boom, GenBank no. KX239026); E & H, v. d. Boom 44999 (hb. v. d. Boom, GenBank no. KX239018); F, v. d. Boom 44837 (hb. v. d. Boom, GenBank no. KX239027). Scales: A, C & E=1 mm; B, D & F=50 μm; G=0·5 mm; H=10 μm. In colour online.
Newly sequenced specimens of B. brandii, represented by two haplotypes that differ in four positions, were collected from widely spread geographical regions of Europe (see Table 1) and form a strongly supported monophyletic clade (100 in both MP and ML methods and 1·00 in BA). Bacidina brandii is most closely related to B. sulphurella (Samp.) M. Hauck & V. Wirth (0·97 in BA); both species are characterized by the dark, Arnoldiana-brown pigmented hypothecium.
Two sequences of B. delicata (Larbal. ex Leight.) V. Wirth & Vězda were downloaded from GenBank for this study. One of these, collected in Sweden (AF282097), seems to be closely related to B. flavoleprosa Czarnota & Guz.-Krzem. but the other, collected in France (JQ796854), seems to be related to B. egenula; both relationships are supported only in BA, 0·93 and 0·99 respectively (Fig. 1).
Discussion
Bacidina mendax sp. nov. is a very variable taxon in its apothecial pigmentation as well as its thallus development; therefore earlier errors in its identification with other representatives of the genus Bacidina are understandable. Apothecia of this species are variously coloured (see Figs 2 & 3) but its conidia are uniformly filiform and often slightly hooked, as in B. sulphurella (Figs 2I & 3F); they are also variable in their length, even within one pycnidium. Molecular variability in B. mendax was also observed: within seven sequenced specimens referred to this species, four ITS haplotypes have been discovered, only three of which formed a poorly supported clade (84 in ML and 0·97 in BA) (Fig. 1). In several specimens of B. mendax, represented by, for example, the designated holotype (Czarnota 4888), the thallus is well developed, thick, and composed of dense, scurfy warts (Fig. 2 A-C &G) with, usually, completely- or semi-immersed pycnidia (Fig. 2H). Other specimens (represented by, for example, Malíček 1769) have a negligible or poorly developed thallus composed of separated, small warts which never form a thick crust (Fig. 3B & C). Since the molecular data we generated are insufficient to support the description of some infraspecific taxa within B. mendax or to show some other phylogenetic relationships, we propose to keep the species in a broad sense. Despite its variability, thallus structure can be used as a key feature in the separation of B. mendax, in a broad sense, from a similar species, B. neosquamulosa (see Figs 2–4), which forms ± coralloid to palmate thallus warts never found in B. mendax.
The new, more complete phylogenetic analysis of the genus Bacidina and additional taxonomic studies presented in this work showed that some representatives of the genus had been determined erroneously. Specimen Czarnota 4888 (GenBank no. JN972444), which was believed to represent B. neosquamulosa (Czarnota & Guzow-Krzemińska Reference Czarnota and Guzow-Krzemińska2012), belongs to a separate lineage regarded as representing the new B. mendax. This specimen has been selected as the holotype of B. mendax. Moreover, as the haplotype of the holotype was found to be identical to the sequence obtained from B. caligans (AF282096) downloaded from GenBank, we suggest that this specimen also represents B. mendax and should be labelled as such (Fig. 1).
Bacidina adastra (GenBank no. JN972442), mentioned in the analysis by Czarnota & Guzow-Krzemińska (Reference Czarnota and Guzow-Krzemińska2012), is nested here within the lineage of B. caligans and renamed as this species. This specimen has small, yellowish punctiform soralia and poorly developed apothecia and was consequently regarded as an immature specimen of B. adastra.
Two sequences of B. delicata downloaded from GenBank are probably not the same species as they are not nested in the same monophyletic clade (Fig. 1). It would appear that at least one of them does not represent this species but the solution to this problem requires further investigation.
The placement of the specimen obtained from GenBank as B. egenula (Ekman 3003, GenBank no. AF282095) is questionable. We sequenced four other specimens of B. egenula and they are not related to the sequence of the GenBank specimen. Our revision of this collection (Norway, Hordaland, Sund, Sotra, Golta, UTM WGS84: 32V KM 787 822, on brick, on the ground, in a pasture, alt. 20 m, 9.3.1997, Ekman 3003, hb. BG, L-65567) showed that it represents another species. It is characterized by short, ±straight, bacilliform to nearly fusiform ascospores, 14–18×2·5–3·5 µm (not acicular and >25 µm in length, as in B. egenula), needle-like, straight or only slightly curved (not sigmoid to strongly curved), 3–5-septate conidia 25–30(–35)×1·5–2·0 µm, small patches of crustaceous, ±irregularly areolate (not granular; without goniocysts) thallus and colourless to slightly yellowish hypothecium (not Arnoldiana-brown pigmented). Such characters may resemble those found in Lecania subfuscula (Nyl.) S. Ekman but comparative studies to confirm this hypothesis have not been carried out.
Analyses of the newly sequenced B. egenula specimens may shed new light on the taxonomy of this species. All sequenced specimens from Poland (Czarnota 8304, 8314 & 8337) have a more or less Arnoldiana-brown pigmented hypothecium characterizing B. egenula, while the Czech collection (Czarnota 5302; see Table 1) has a completely colourless hypothecium and is therefore tentatively named Bacidia viridescens (A. Massal.) Norman since all its characters correspond well with the widely accepted description of this species (e.g. Coppins & Aptroot Reference Coppins and Aptroot2009; Wirth et al. Reference Wirth, Hauck and Schultz2013). The hypothecium pigmentation usually differs between these two species (Coppins & Aptroot Reference Coppins and Aptroot2009). Considering this result, the real relationship between Bacidina egenula and Bacidia viridescens remains unclear and should be resolved in further phylogenetic studies based on additional samples of specimens thought to be B. viridescens.
One specimen of Bacidina brandii collected in Poland was used for the phylogenetic analyses. Rejecting the erroneously published previous record (Czarnota Reference Czarnota2016), the specimen presented here confirms the occurrence of this species in Poland and the Carpathians.
Our analyses showed that species belonging to the genus Bacidina require further study. Some relationships within the genus are still unresolved and some species, such as B. delicata, should be analyzed in greater detail.
Taxonomy
Bacidina mendax Czarnota & Guz.-Krzem. sp. nov.
MycoBank No.: MB 821694
Differing from B. neosquamulosa in the lack of a subsquamulose thallus and from B. caligans in its longer and only slightly curved to apically hooked conidia and lack of granular (sorediate) thallus.
Type: Slovakia, Považský Inovec, Tematíske Kopce, between Lúka Village and Tematísky hrad Castle, 48°39'52''N, 17°53'56''E, alt. c. 230 m, on roadside Acer platanoides, 22 April 2006, P. Czarnota 4888 (GPN—holotype; UGDA—isotype).
Thallus inconspicuous to distinct, crustaceous, straw-coloured to bright green, composed of minute, merged granules to form a thin, scurfy, uneven crust or small warts. Photobiont chlorococcoid, with large, globose cells up to 17 µm diam.
Apothecia 0·2–0·7 mm diam., constricted at the base, variable in colour, from whitish and flesh-coloured throughout, beige, pinkish-buff, brownish to grey-brown and dark fuscous-brown, marginate at the beginning, later sometimes with margin excluded; disc concolorous, paler or darker than the margin; in the case of less coloured apothecia, margin usually at least partially extraneously dark-pigmented. Excipulum (30–)40–60(–70) µm wide, composed of radiating hyphae 1·5–2·5 µm in width, but 1–2 rows of cells of outermost part with lumina widening to 5–7 µm; upper and outer part of excipulum pinkish orange, brown, fuscous brown and then K– or K± pinkish to purplish or olive-brown, K± intensifying; inner part of excipulum colourless to, ±straw-coloured towards the hypothecium and below hypothecium sometimes paraplectenchymatous, composed of ±globose to elongate cells of 3–5(–7) µm wide lumina. Hypothecium to 60 µm tall, colourless to ±straw-coloured. Hymenium (40–)50–60(–65) µm tall, colourless or sometimes with ±pinkish to olive-brown, K− or K+ intensifying epihymenium as well as vertical streaks; pigment confined to walls of paraphyses and asci. Paraphyses of two types: first, simple, 1·5–2·0 µm wide, straight to slightly curved with globose apical cells, slightly widening to 2·5(–3·0) µm; second, irregular, stout, 3–4 µm wide, sometimes forming vertical streaks, forked with pigmented globose to elongated apical cells, up to 6 µm in width. Asci cylindrical to narrowly clavate, 7–9×(25–)35–40(–45) µm, Lecanora-type. Ascospores acicular, (25–)30–38(–40)×1·2–1·5 µm, 3–5(–6)-septate, straight.
Pycnidia pale-coloured throughout, globose, ±immersed within the thallus warts, 0·15–0·30 mm diam. Conidia filiform, ±straight to slightly curved (usually in upper half) or ±hooked, (25–)35–55(–62)×1·0–1·2 µm, 3–6-septate.
Chemistry. No substances detected by TLC.
Etymology. ‘Mendax’, from its similarity with other related species.
Remarks. A provisional revision of material stored in several European herbaria showed that Bacidina mendax had already been collected in the past, but its darker morphs with a poorly developed thallus were usually identified as B. caligans, while collections forming a thicker, warted thallus were identified as B. neosquamulosa. Specimens with predominantly pale-coloured apothecia were also misidentified as B. phacodes or B. delicata. Indeed, morphological variation in B. mendax can be a source of taxonomic error, particularly since in the same collection pale brown to fuscous brown apothecia can grow together or, depending on the circumstances, can be almost entirely pale or dark brown. This extensive apothecial variability also determines the internal pigmentation which is either very pale brown throughout or distinctly dark brown in the outermost part of the excipulum and hymenium and being confined to walls of excipular hyphae, apical parts of paraphyses or asci. The type of pigmentation does not seem to be a stable feature since it can often be represented by the fuscous brown, K± purplish pigment or by grey-brown, brown to orange-brown, K+ intensifying or K− pigments, or a mixture of these pigments. For a detailed comparison of the main diagnostic features of B. mendax and several European representatives of the genus resembling different morphs of this species, see Table 2 and Figs 2–5.
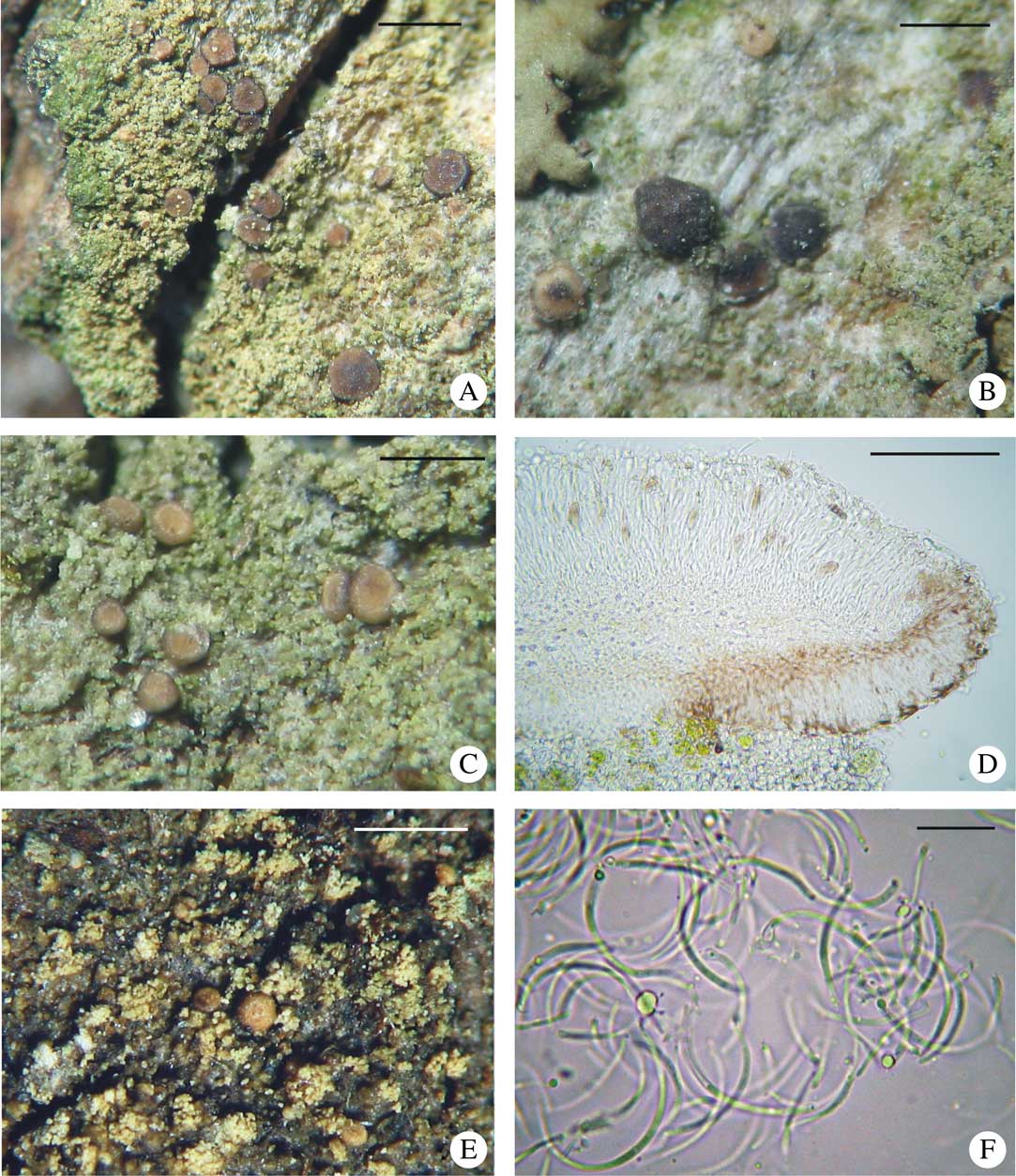
Fig. 5 Bacidina caligans. A–C & E, habitus of various morphotypes; D, section through apothecium; F, pycnospores. A–D, v. d. Boom 44973 (hb. v. d. Boom, GenBank no. KX239024); E & F, Czarnota 4961 (GPN, GenBank no. JN972442). Scales: A–C & E=1 mm; D=100 μm; F=10 μm. In colour online.
Table 2 A comparison of main diagnostic characters for Bacidina mendaxand similar European corticolous representatives of the genus with colourless hypothecium

Distribution and habitat. The new Bacidina species, reported here from several European regions, is probably widespread in the central-eastern part of the continent. It is a tolerant epiphyte found on deciduous trees and shrubs (Acer, Betula, Carpinus, Fraxinus, Malus, Populus, Pyrus, Quercus, Salix, Sambucus and Tilia), usually in mostly anthropogenic habitats including roadside trees in large cities, urban parks and on riversides, and solitary trees in rural landscapes, orchards and edges of woodland, as well as in natural, old-growth forests (such as strictly protected areas of the Białowieża National Park). Bacidina mendax occurs on trunks, twigs and branches of trees, usually in association with many lichens tolerant of both air pollution and nitrification (e.g. Amandinea punctata, Lecania naegelii, Physcia adscendens, P. tenella, Scoliciosporum spp. and Xanthoria parietina). On the other hand, in natural woodlands B. mendax was found to grow in association with some old-growth forest indicators such as Biatora hemipolia, Bacidia laurocerasi, B. subincompta and Fellhanera gyrophorica.
Additional specimens examined. Czech Republic: Rychlebské hory Mts: c. 0·5 km W of Bíla Voda Village, limestone quarry Kukačka, 50°26'29''N, 16°53'01''E, alt. c. 350 m, on Salix sp., 2004, M. Kukwa 3140 (UGDA-L-10926). Středočeská pahorkatina foothills, ‘Husova kazatelna’ hill, c. 1 km NE of Petrovice Village, 49°34'03''N, 14°21'49''E, alt. 500 m, on Betula pendula, 2008, P. Czarnota 5327 (GPN); Praha – Řepy, Hekova Street in front of supermarket ‘Plus’, 50°04·28'N, 14°18·18'E, alt. 350 m, on pollarded Acer platanoides, 2007, Z. Palice 11110 & J. Palicová (PRA). S Moravia: Břeclav District, Lanžhot, Cahnov-Soutok National Nature Reserve, old-growth forest in flood plain, 7·5 km SSW of Břeclav Town, 48°39'09''N, 16°56'20''E, alt. 150 m, on log of Fraxinus within old-growth deciduous forest, 2014, P. Czarnota 7571 (GPN).—Great Britain: England: V.C. 17, Surrey, London, Kew, Royal Botanic Garden, on Quercus sp., 2005, M. Kukwa 4568 (UGDA-L-12931).—Poland: Pomerania: Mierzeja Wiślana sandbar, Sobieszewska Island, 1·5 km N of Świbno by Vistula River, 54°20'42''N, 18°56'23''E, on riverside Salix sp., 2006, M. Kukwa 5171 (UGDA-L-13408); Mierzeja Wiślana sandbar, between Piaski Village and Krynica Morska Town, forest by Zalew Wiślany Bay, 54°24'51''N, 19°33'41''E, on Salix sp., 2004, M. Kukwa 2966 (UGDA-L-11718); on Sambucus nigra, 2004, M. Kukwa 2960 (UGDA-L-11712). Iławskie Lakeland: Postolińska Struga Valley, c. 0·5 km NNW of Nowa Wieś Village, on twigs of Salix sp., 2004, M. Kukwa 3611 (UGDA-L-12339); ibid., c. 1 km NE of Mątki Village, on Salix sp. in a gravel pit, 2001, M. Kukwa 99 (UGDA-L-12514). Fordońska Valley: near Kiełp Village, close to ‘Zbocza Płutowskie’ Nature Reserve, 53°17'28''N, 18°22'50''E, on roadside Salix sp., 2004, M. Kukwa 3512 (UGDA-L-11187). Wysoczyzna Ciechanowska plateau, Gąsocin Village, 52°44'38·5''N, 20°42'42·1''E, on Sambucus nigra at the edge of alder forest, 26 xii 2009, D. Kubiak (OLTC-L-3397 & 3398). Równina Bielska plain, Białowieża Primeval Forest, Białowieża National Park, forest section no. 256, 52°46'28''N, 23°51'37''E, on young Carpinus betulus in Carici elongatae-Alnetum community, viii 2014, M. Kukwa & A. Łubek s. n. (KTC; loc. P1); ibid., 52°46'15''N, 23°51'58''E, on young Quercus sp., viii 2014, M. Kukwa & A. Łubek s. n. (KTC; loc. J5); ibid., 52°46'18''N, 23°52'30''E, on Fraxinus excelsior in Circeo-Alnetum community, viii 2014, M. Kukwa & A. Łubek s. n. (KTC; loc. M11). Beskid Mały Mts: Kocierz Rychwałdzki Village, alt. 500 m, on roadside Populus sp., 10 v 1962, J. Nowak (KRAM-L-9455). Kotlina Jasielsko-Krośnieńska basin, Podole Village by the Wisłok River, 49°38'37·3''N, 21°54'43·0''E, on riverside Salix fragilis, 2010, P. Czarnota 7244 (GPN). Beskid Niski Mts: Magurski National Park, forest section no. 158, close to Nieznajowa settlement, by Wisłoka River, on riverside Salix alba, alt. 435 m, 11 ix 2009, E. Adamska, D. Bielec, J. Kozik & A. Łubek (KRAP).—Slovakia: Považský Inovec: Tematíske Kopce, between Lúka Village and Tematísky hrad Castle, 48°39'52''N, 17°53'56''E, alt. c. 230 m, on roadside Acer platanoides, 2006, M. Kukwa 4992 (UGDA-L-13227—topotype). S Moravia: near Morava River, Horny les National Nature Reserve, 48°21'09''N, 16°51'47''E, alt. 150 m, on Quercus robur branch in old-growth forest in flood plain, 2014, P. Czarnota 7786 (GPN).—Ukraine: Eastern Carpathians: Ivano-Frankivsk Region, Rozhniativ District, 1·5 km W of Ivanivka Village, 48°53'09·8''N, 24°05'29·8'', alt. 490 m, on Quercus sp. in young oak-hornbeam forest, a former wood pasture, 2015, P. Czarnota 8117, 8119, 8157, 8158, 8159 & 8160 (GPN).
New combination
Bacidina pycnidiata (Czarnota & Coppins) Czarnota & Guz.-Krzem. comb. nov.
MycoBank No. MB 821695
Basionym: Bacidia pycnidiata Czarnota & Coppins, Lichenologist 38: 407–408 (2006); type: Czech Republic, Eastern Sudetes, Rychlebské hory Mts, W of Bila Voda Village, vicinity of worked-out marble quarry ‘Kukačka’ near the border of Poland, 50°26'18''N, 16°53'14''E, alt. c. 360 m, on bryophytes over marble rock within mixed spruce-ash forest, 23 April 2004, P. Czarnota 4157 (GPN—holotype; E, UGDA—isotypes).
Remarks. Owing to the placement of Bacidia pycnidiata within the monophyletic group of species that correspond by their characters to the conserved genus Bacidina (Ekman 1996 Reference Ekmanb ) (Fig. 1), B. pycnidiata is transferred to Bacidina. Despite the nomenclatural problem related to a choice of Bacidia phacodes Körb. as the type species for the genus Bacidina (Vězda Reference Vězda1990), it is at least clear now that B. pycnidiata is not nested within Bacidia s. str.
The first author thanks the Polish National Science Centre for partial support of this research, grant no. NCN 2013/11/B/NZ9/00793. The study was also supported by the University of Gdańsk task grant no. 530-L140-D242-14. We are grateful to the curator of the lichen herbarium BG (University Museum of Bergen), Tor Tønsberg, for making available the sequenced B. egenula specimen (Ekman 3003; GenBank no. AF282095) for our revision, and to Martin Kukwa, Jiří Malíček, Zdeněk Palice, Pieter van den Boom, Darek Kubiak and Anna Łubek for the opportunity to use their collections in this work. Finally, we warmly thank Mark Seaward for linguistic improvements and anonymous reviewers for all constructive corrections and valuable comments.









