Introduction
The yak, known in China as the ‘almighty livestock’, is well adapted to low temperatures, low oxygen levels, and the low pressure environment found at high altitudes (e.g. at 3500 m above sea level). These animals are able to make full use of alpine grasslands, which other livestock find difficult to utilize. However, this environment provides primitive grazing with vegetation that grows slowly, resulting in low animal production yields, in addition the deteriorating environment makes yak populations vulnerable to environmental pressures. How to accelerate the genetic breeding of yaks, thereby not making them vulnerable to extinction, and to increase the value of their production is an important problem with a worldwide focus.
Much progress has been made towards applying assisted breeding to aid yak conservation, such as hybridization between cattle and yak species. In particular, interspecies somatic cell nuclear transfer (iSCNT) is a novel method that has been shown to protect species that are not subject to genetic variation and are in danger of extinction. The sand cat (Gómez et al., Reference Gómez, Pope, Kutner, Ricks, Lyons, Ruhe, Dumas, Lyons, López, Dresser and Reiser2008), gaur (Vogel Reference Vogel2001), mouflon (Loi et al., Reference Loi, Ptak, Barboni, Fulka, Cappai and Clinton2001) and grey wolf (Oh et al., Reference Oh, Kim, Jang, Kim, Hong, Park, Park, Park, Sohn, Kimb, Shin and Lee2008) have all been cloned successfully using iSCNT, although the efficiency is still extremely low. The efficiency of iSCNT depends on a variety of factors, such as the status of donor cells, the quality of recipient oocytes and the environment of the in vitro operation. However, the low success rate found with this technique has been widely attributed to incomplete reprogramming of epigenetic modifications or to epigenetic errors (Couldrey & Lee, Reference Couldrey and Lee2010).
It has been reported that histone acetylase levels in cloned SCNT embryos of intraspecies bovine, mouse, pig, sheep and rabbit that had been treated with histone deacetylases inhibitors (HDACi), such as TSA and VPA, were improved significantly compared with untreated animals and were similar to their in vitro fertilization (IVF) or in vivo counterparts (Lager et al., Reference Lager, Ragina, Ross, Beyhan, Cunniff, Rodriguez and Cibelli2008). Furthermore these embryos showed improved development to the blastocyst stage (Beebe et al., Reference Beebe, Mcllfatrick and Nottle2009; Shao et al., Reference Shao, Ding, Gao, Ding, Gao, Li, Wu, Xu and Liu2009; Costa-Borges et al., Reference Costa-Borges, Santalo and Ibanez2010; Zhao et al., Reference Zhao, Hao, Ross, Spate, Walters, Samuel, Clifton and Randall2010), plus there was an increase in the number of live offspring after transfer to foster mothers (Maalouf et al., Reference Maalouf, Liu, Brochard, Renard, Debey, Beaujean and Zink2009; Costa-Borges et al., Reference Costa-Borges, Santalo and Ibanez2010), and reduced abnormal phenotypes, with the exception of placental overgrowth in mice (Kishigami et al., Reference Kishigami, Mizutani, Ohta, Takafusa, Thuan, Wakayama, Bui and Wakayama2006). Even though improvements in cloning efficiency have been observed in intraspecies cloned embryos with TSA treatment, conflicting results have been reported when interspecies cloned embryos were treated with TSA (Shi et al., Reference Shi, Miao, Ouyang, Huang, Lei, Yang, Han, Song, Sun and Chen2008; Srirattana et al., Reference Srirattana, Laowtammathron, Devahudi, Imsoonthornruksa, Sangmalee, Tunwattana, Lorthongpanich, Sripunya, Keawmungkun, Phewsoi, Ketudat-Cairns and Parnpai2008).
To our knowledge, the effect of pre-treatment of yak iSCNT embryos with NaBu has not been reported previously and the effects of NaBu on the development competence and expression patterns of genes related to development in yak iSCNT embryos after activation have not been thoroughly examined. In the present study, we examined whether treatment of yak iSCNT embryos with NaBu improved developmental competence in vitro and ‘corrected’ expression patterns of genes (Oct-4, HDAC-2, Dnmt-1, and IGF-1) during development stages in vitro.
Materials and methods
All chemicals used in this study were purchased from Sigma (St. Louis, MO, USA) unless otherwise noted. Disposable, sterile plasticware were purchased from Nunclon (Roskilde, Denmark).
All procedures in this experiment were approved by the Animal Care and Use Committee of Southwest University for Nationalities (Chengdu, China) and performed in accordance with animal welfare and ethics.
Nuclear donor cell preparation
Yak fibroblast cells from the ear skin were derived as described previously (Xiong et al., Reference Xiong, Wang, Zi, Ma, Xu, Wang and Li2012). Briefly, the tissues were minced into pieces (1 mm3) using sterile scissors in a 35 mm Petri dish, then explants were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 1 mM sodium pyruvate, 1 μg/ml EGF, 100 IU/ml penicillin and 100 mg/ml streptomycin under 5.5% CO2 in air at 38.5°C. The cells were trypsinized and reconstituted at a concentration of 1 × 106 cells/ml after reaching 80–90% confluency. Then 2–5 cells were passaged, as donor cells for iSCNT, and were synchronized in the G0/G1 phase by contact inhibition for 2 days before iSCNT.
In vitro maturation (IVM)
Bovine ovaries were obtained from the local slaughter houses and transported to the laboratory within 4 h of slaughter in sterile 0.9% (w/v) NaCl saline at approximately 20°C in a thermos bottle. Cumulus–oocyte complexes (COCs) were aspired from 2–8 mm follicles using a 12-gauge needle and then injected into phosphate-buffered saline (PBS) plus 0.5 IU/ml heparin and 5% (v/v) FBS. COCs showing an even cytoplasm and surrounded by compact cumulus cells were collected from the follicular fluid and PBS mixture, washed twice in PBS and then incubated in TCM-199 (Gibco) supplemented with 10% (v/v) FBS, 1 μg/ml 17β-estradiol and 0.075 IU/ml human menopausal gonadotropin for 20 h at 38.5°C in 5.5% CO2 in air.
Parthenogenetic activation (PA)
PA was conducted according to the method described previously by Xu & Yang (Reference Xu and Yang2001). The matured oocytes were denuded of cumulus cells in PBS supplemented with 0.2% (v/v) hyaluronidase and activated in 5 μM ionomycin for 5 min, followed by a 4 h exposure to 2 mM dimethylaminopurine (6-DMAP) in mSOF. The activated oocytes were washed twice in mSOF and then randomized into five groups, and cultured in mSOF plus either 0 nm (control), 5 nm or 10 nm NaBu for 6 h and 12 h, respectively. After culture with NaBu, the embryos were transferred to NaBu-free mSOF for further culture at 38.5°C in 5.5% CO2 in air for 7 days.
Interspecies somatic cell nuclear transfer (iSCNT)
iSCNT was conducted according to the method described previously by Xiong et al. (Reference Xiong, Wang, Zi, Ma, Xu, Wang and Li2012). Briefly, after IVM, matured bovine oocytes were denuded with 0.2% (v/v) hyaluronidase in PBS to disperse the cumulus cells. Enucleation was performed using a 20-μm (internal diameter) glass pipette and by aspirating the first polar body and a small amount of the surrounding cytoplasm. After enucleation, a single synchronized yak fibroblast cell was introduced into the perivitelline space of the enucleated oocyte and fusion was induced by application of two electrical pulses of 35 V for 10 μs. Reconstructed embryos were stored in mSOF that contained 5 g/l cytochalasin B for 2 h until activation. Then, successfully reconstructed embryos were activated in 5 mM ionomycin for 5 min followed by 4 h of exposure to 2 mM 6-dimethylamino-pyridine (6-DMAP) in mSOF. Activated embryos were cultured in mSOF in randomized groups. After culture in mSOF with NaBu for different lengths of time, embryos were transferred to NaBu-free mSOF for further culture at 38.5°C in 5.5% CO2 in air for 7 days.
In vitro fertilization (IVF)
Frozen–thawed yak semen was thawed, and motile spermatozoa were obtained for fertilization using a Percoll gradient (Pharmacia, diluted into 45% and 90%). Viable sperm were recovered and resuspended. Maturated oocytes were co-incubated with spermatozoa at a concentration of 1 × 106 cells/ml for approximately 24 h in a humidified atmosphere with 5.5% CO2 in air at 38.5°C. Then bovine–yak IVF (BY-IVF) presumptive zygotes were denuded by treatment with 0.2% (v/v) hyaluronidase in PBS, washed twice in mSOF, and then transferred into mSOF for further culture as yak iSCNT embryos.
Counting of cell numbers
Day 7 blastocysts from BY-IVF and iSCNT were permeabilized in 4% paraformaldehyde in PBS for 20 min, stained with 10 μg/ml propidium iodide (PI) for 15 min, and then mounted on slides in 5 μl glycerol. At least 10 embryos from each group were selected randomly for processing. The total number of cells was counted under an epifluorescence microscope (Nikon, Japan) using a digital camera.
Real-time reverse transcription polymerase chain reaction (real-time RT-PCR)
mRNA was extracted using the RNeasy total extraction kit (Qiagen, Valencia, USA) in accordance with the manufacturer's protocol but with some modifications. Briefly, two embryos from BY-IVF, iSCNT and iSCNT-NaBu groups were chosen randomly, washed twice in Ca2+- and Mg2+-free PBS, and 10 μl lysis buffer was added for 3 min. cDNA synthesis was achieved using the cDNA synthesis kit (Takara, China) in accordance with the manufacturer's instructions.
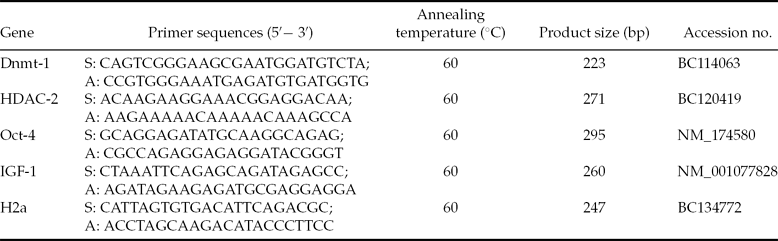
Real-time RT-PCR was performed using the five primer sets shown in Table 1, primers were designed using the Primer 5.0 software. The specificities of all primers were tested using BLAST analysis against the genomic National Center for Biotechnology Information, USA (NCBI) database, and PCR products were sequenced for verification.
Table 1 Primer sequences and PCR conditions used for real-time PCR

S: forward primer; A: reverse primer.
Quantitative real-time PCR (qRT-PCR)
All qRT-PCR reactions were performed in triplicate in a 20-μl reaction volume using the quantitative real-time PCR CFX96 detection system (Bio-Rad, Hercules, USA) and using reaction mixture SYBR Premix Ex Taq™ II (TaKaRa, China) that contained 10 μl 2× SYBR Green premix, plus 0.8 μl of forward and reverse primers (20 pmol/ml), 2 μl embryonic cDNA and 6.4 μl RNase- and DNase-free water. Histone H2a was used initially as the housekeeping reference gene. All primers used were optimized to ensure similar reaction efficiencies (96–98%) between target genes and H2a. Thermal cycling conditions were 95°C for 10 min, followed by 40 PCR cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. The melting protocol was a step cycle starting at 65°C, and then increased to 95°C with 0.5°C/5 s increments. In addition, a non-template control that contained the above reaction mixture but lacking cDNA was included in each PCR run as a negative control. The relative quantification of gene expression levels was conducted using the 2−ΔΔCt method.
Statistical analysis
The experiment was repeated at least three times for each treatment group. Data are presented as mean ± standard error of the mean (SEM). The 2-cell stage embryo, 8-cell stage embryo, and blastocyst formation, total cell number and the gene expression levels among those groups were tested by one-way analysis of variance (ANOVA) and least significant difference (LSD) test using the SPSS 17.0 software. Differences were considered significant at P-values< 0.05.
Results
Effects of NaBu on the in vitro development of PA embryos
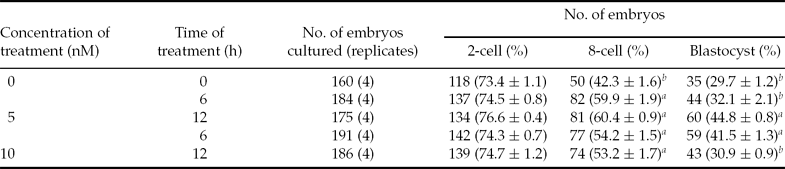
First, the effect of NaBu on the in vitro development of PA embryos was evaluated. In this study, parthenotes were chosen because they were relatively easy to produce in large numbers for the analysis, and the chemical activation protocol of PA embryos was similar to that of iSCNT embryos. NaBu concentrations of 1–10 nM and the treatment times of 4–20 h have been reported in different species (Shi et al., Reference Shi, Miao, Ouyang, Huang, Lei, Yang, Han, Song, Sun and Chen2008). Parthenotes were exposed to different concentrations of NaBu (0, 5, 10 nM) for 6 h or 12 h, respectively, then cultured in vitro for 7 days. As shown in Table 2, NaBu did not have any negative effect on cleavage rates (P> 0.05), but there was a significant beneficial effect on 8-cell stage and blastocyst formation rates when PA embryos were treated with 5 nM NaBu for 12 h. However, with the increase in treatment concentration and exposure time, the blastocyst formation rate was in a downward trend that might be due to the toxicity of NaBu. Therefore, a 12 h treatment period plus a 5 nM NaBu concentration was chosen for treating iSCNT embryos.
Table 2 Effects of treated concentration and expose time of NaBu on the in vitro development of parthenotes embryos

a ,b Values with different superscripts within a column are significantly different (P < 0.05).
2-cell rate: no. of 2-cell stage embryos/no. of embryos cultured. 8-cell rate: no. of 8-cell stage embryos/no. of 2-cell stage embryos. Blastocyst rate: no. of blastocysts/no. of 2-cell stage embryos.
Effects of NaBu on the in vitro development of yak iSCNT embryos
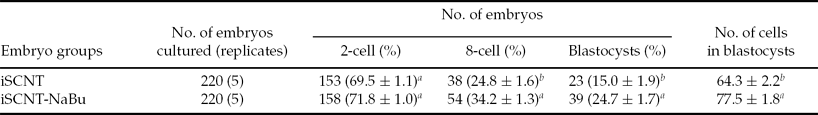
The yak iSCNT embryos were divided randomly into two groups and treated with 5 nM NaBu for 12 h after activation, and then the in vitro development and quality of yak iSCNTs were evaluated. As shown in Table 3, although there was no significant difference on the rate of cleavage after NaBu treatment (P> 0.05), this treatment significantly improved the rate of 8-cell stage embryo (34.2 ± 1.3% vs. 24.8 ± 1.6%, P< 0.05) and blastocyst formation (24.7 ± 1.7% vs. 15.0 ± 1.9%, P< 0.05) compared with their controls; the treatment of NaBu had a positive effect on the total cell number per blastocyst (shown in Fig. 1).
Table 3 Effects of NaBu on the in vitro development of yak iSCNT embryos

a ,b Values with different superscripts within a column are significantly different (P < 0.05).
2-cell rate: no. of 2-cell stage embryos/no. of embryos cultured. 8-cell rate: no. of 8-cell stage embryos/no. of 2-cell stage embryos. Blastocyst rate: no. of blastocysts/no. of 2-cell stage embryos.

Figure 1 The total cell number of blastocysts derived from interspecies somatic cell nuclear transfer (iSCNT) (A), iSCNT-NaBu (B) and bovine–yak in vitro fertilization (BY-IVF) (C) (×200 magnification).
Effects of NaBu on gene expression of yak iSCNT embryos
The relative transcript abundance of Oct-4, HDAC-2, Dnmt-1 and IGF-1 was quantified by real-time PCR at various preimplantation developmental stages of BY-IVF and yak iSCNT embryos with or without NaBu treatment to examine if the low cloning efficiency was correlated with gene expression during development in vitro. Figure 2 shows that the relative expression level of Oct-4 in yak iSCNT embryos was significant lower compared with that of their BY-IVF counterparts after the 2-cell stage; after NaBu treatment, the expression levels of Oct4 increased significantly in yak iSCNT embryos, which exhibited similar levels to their BY-IVF counterparts. The expression patterns of HDAC-2 and IGF-1 were observed in yak iSCNT and BY-IVF embryos, the NaBu-treated iSCNT embryos showed lower levels of expression for HDAC-2 and IGF-1, and these levels were more similar to their BY-IVF counterparts. The level of DNA methylation related gene, Dnmt-1, was higher in iSCNT embryos than in BY-IVF counterparts, and there was no significant difference between iSCNT and iSCNT-NaBu groups up to the blastocyst stage.

Figure 2 Relative transcript abundance of Oct-4, HDAC-2, Dnmt-1 and IGF-1 genes in embryos produced by bovine–yak in vitro fertilization (BY-IVF), interspecies somatic cell nuclear transfer (iSCNT) or iSCNT-NaBu at four stages of embryonic development (2-cell stage embryo, 8-cell stage embryo, morula, and blastocyst). Different letters denote samples that differed significantly within each developmental stage (P< 0.05).
Discussion
Global epigenetic reprogramming has been reported as a major factor that is required to take place following SCNT for normal development and successful cloning. The epigenetic state of the donor nucleus must be erasure and an embryonic epigenetic state pattern should be established in SCNT embryos. Although the mechanism of how epigenetic states modify a donor cell after iSCNT is still a mystery, several studies have shown that nuclear–cytoplasmic incompatibilities between species were the major handicaps for iSCNT (Martha et al., Reference Martha, Gomez, Earle, Monica, Biancardi, Galiguis, Morris, Wang and Dresser2011).
There is growing evidence that suggests that proper use of histone deacetylase inhibitor (HDACi) can improve the efficiency of SCNT. Lee et al. (Reference Lee, Yu, Bang, Cho, Gautam, Kim and Kong2010) reported that treating cat cells with TSA before iSCNT, significantly increased the level of acetylation in histone H3K9, and improved the in vitro developmental competence and total cell numbers of iSCNT blastocysts. Our previous study showed that SCNT embryos pre-treated with TSA and Scriptaid significantly improved the efficiency of bovine cloning (Wang et al., Reference Wang, Zhang, Wang, Xu, Xiong, Li, Su, Hua and Zhang2011). Likewise we observed that in iSCNT yak embryos treated with NaBu, a novel HDACi substantially increased cloned embryo development in vitro. Our results are consistent with those of Das et al. (Reference Das, Gupta, Uhm and Lee2010) and of Mohana et al. (Reference Mohana, Song, Cho, Balasubramanian, Choe and Rho2007), who both found that NaBu can increase the blastocyst formation rate in SCNT embryos. However, a discrepancy exists in its effectiveness in donor cells. Das et al. (Reference Das, Gupta, Uhm and Lee2010) confirmed that there was no effect on cloning efficiency when donor cells were pre-treated with NaBu. This finding was in sharp contrast with the report by Shi et al. (Reference Shi, Hoeflich, Flaswinkel, Stojkovic, Wolf and Zakhartchenko2003). This discrepancy may be due to species differences in response to NaBu in these studies. In our study, we also found no positive effect when donor cells were pre-treated with NaBu (data not show). Interestingly, we discovered that most yak iSCNT embryos were arrested at the 2- to 8-cell stage, and that the 8-cell stage embryos increased predominantly when yak iSCNT embryos were treated with 5 nM NaBu for 12 h after activation. Therefore, we concluded that the lower levels of development in vitro of iSCNT yak embryos may be related to aberrant gene expression at this stage, and that NaBu could ‘correct’ the aberrant gene expression of iSCNT yak embryos during development in vitro, especially during the maternal zygotic transition (MZT) stage.
The onset of transcription in embryonic genome activation is one of the most critical events of early embryogenesis. However, previous studies have indicated that the main reason for low efficiency in iSCNT was because correct transcription failed to occur (Bui et al., Reference Bui, Wakayama, Kishigami, Park, Kim, Thuan and Wakayama2010). This transcriptional activation is a remarkable event, during which a number of genes are activated and there is dramatic reprogramming of embryonic gene expression, including those genes that are important for successful embryo development. Consequently, we determined expression patterns of Oct-4, HDAC-2, Dnmt-1 and IGF-1 that may in part regulate gene expression during early embryogenesis of yak iSCNT embryos. Oct-4 is a key pluripotency specific gene for which accurate expression is crucial for preimplantation embryo development; reactivation of Oct-4 expression is a marker of nuclear reprogramming (Miyamoto et al., Reference Miyamoto, Furusawa, Ohnuki, Goel, Tokunaga, Minami, Yamada, Ohsumi and Imai2007). HDAC-2 is a member of the histone deacetylases, and participates in removal of acetyl moieties from histone tails (Murko et al., Reference Murko, Lagger, Steiner, Seiser, Schoefer and Pusch2010). Dnmt-1, a DNA methyltransferases, is expressed constitutively and is responsible for maintenance of global methylation following DNA replication (Bosak et al., Reference Bosak, Fujisaki, Kiuchi, Hiraiwa and Yasue2003); Deletion of Dnmt-1 alone results in embryonic lethality (Okano et al., Reference Okano, Bell, Haber and Li1999). Insulin-like growth factor 1 (IGF-1) is a classic imprinted gene that plays a crucial role in embryo development and viability (Velazquez et al., Reference Velazquez, Zaraza, Oropeza, Webb and Niemann2009). Dramatic increase in Oct-4 expression levels was observed after iSCNT embryos were treated with NaBu, along with significantly decreased levels of HDAC-2, Dnmt-1 and IGF-1 that were more similar to their BY-IVF counterparts. These results confirmed our hypothesis that NaBu ‘corrected’ the expression patterns of developmentally important genes in early development of yak iSCNT embryos, and these data were consistent with previous studies (Li et al., Reference Li, Kato, Tsuji and Tsunoda2008; Martha et al., Reference Martha, Gomez, Earle, Monica, Biancardi, Galiguis, Morris, Wang and Dresser2011; Ning et al., Reference Ning, Li, Liang, Yang, Xu, Lu, Lu and Lu2012), all of these reports found that gene expression patterns in cloned embryos were normalized after treatment with HDACi. Abnormal expression patterns in crucial genes at the early developmental stage might explain the block in development observed in the present study between the 2-cell and 8-cell stages. Interestingly, the relative expression level of IGF-1 was higher in iSCNT yak embryos compared with their BY-IVF counterparts, especially at the 2-cell stage. Therefore, we propose that IGF-1 can be used as a marker for evaluating the developmental potential of preimplantation iSCNT embryos.
In addition, the 8-cell stage rate, blastocyst formation rate and total cell number in blastocyst of iSCNT embryos were lower compared with their IVF counterparts, and increased significantly after NaBu treatment. This finding indicates that the addition of NaBu improved embryo quantity and quality in vitro. NaBu may alter the activity of histone deacetylases, thus changing the epigenetic model and gene expression patterns, to promote the transition of maternal genes to embryo genes and reduce the block in yak iSCNT embryo development. These findings are in contrast with those of Martha et al. (Reference Martha, Gomez, Earle, Monica, Biancardi, Galiguis, Morris, Wang and Dresser2011) who confirmed that HDACi did not improve the viability of in vitro or in vivo cloned cat embryos. Different HDACi, treatment time and concentration might be the main reason for this discrepancy. However, future research needs to be carried out to further explore the mechanisms of this phenomenon, and to evaluate the long-term effects of NaBu on the developmental competence of yak iSCNT embryos in vivo.
In conclusion, we have demonstrated that NaBu induced improvements in cloning efficiency and gene expression levels at the early developmental stages of yak iSCNT embryos. However, details of the mechanism of nuclear reprogramming of NaBu-treated iSCNT yak embryos are still unclear and need further investigation. On the basis of our results, we propose that the inability of bovine cytoplasm to modify the epigeneticity of yak nuclei after iSCNT resulted in abnormal gene expression during preimplantation development of cloned embryos in vitro, and significantly affected epigenetic reprogramming.
Acknowledgements
The present study was supported by the National Science and Technology Program of China (No. 2012BAD13B06) and the Fundamental Research Funds for the Central Universities of Southwest University for Nationalities (No. 13NZYQN24).
Author disclosure statement
The authors declare that no conflicting financial interests exist.