Introduction
A wealth of data suggests that exposure to an adverse maternal environment in prenatal life, such as obstetric complications, malnutrition, depressed or anxious mood, and maternal treatment with synthetic glucocorticoids, may exert adverse consequences upon fetal brain development thereby altering neurobehavioral trajectories and increasing risk of neuropsychiatric disorders in later life (Roseboom et al. Reference Roseboom, de Rooij and Painter2006; Cottrell & Seckl, Reference Cottrell and Seckl2009; Pesonen et al. Reference Pesonen, Raikkonen, Lano, Peltoniemi, Hallman and Kari2009; Räikkönen et al. Reference Räikkönen, Seckl, Heinonen, Pyhälä, Feldt, Jones, Phillips, Pesonen, Matthews, Lahti, Eriksson, Järvenpää, Strandberg and Kajantie2010; Glover, Reference Glover2011; Tuovinen et al. Reference Tuovinen, Räikkönen, Kajantie, Henriksson, Leskinen, Pesonen, Heinonen, Lahti, Pyhälä, Alastalo, Lahti, Osmond, Barker and Eriksson2012; Reynolds et al. Reference Reynolds, Labad, Buss, Ghaemmaghami and Räikkönen2013). While the biological mechanisms underpinning this developmental plasticity phenomenon, dubbed ‘programming’, still remain elusive, data from experimental animal (Edwards et al. Reference Edwards, Benediktsson, Lindsay and Seckl1993; Lindsay et al. Reference Lindsay, Lindsay, Waddell and Seckl1996a , Reference Lindsay, Lindsay, Edwards and Seckl b ; Benediktsson et al. Reference Benediktsson, Calder, Edwards and Seckl1997; Seckl, Reference Seckl1998; Welberg et al. Reference Welberg, Seckl and Holmes2000; Liu et al. Reference Liu, Li and Matthews2001; Moss et al. Reference Moss, Sloboda, Gurrin, Harding, Challis and Newnham2001; Seckl & Meaney, Reference Seckl and Meaney2004; Holmes et al. Reference Holmes, Abrahamsen, French, Paterson, Mullins and Seckl2006; de Vries et al. Reference de Vries, Holmes, Heijnis, Seier, Heerden, Louw, Wolfe-Coote, Meaney, Levitt and Seckl2007; Seckl & Holmes, Reference Seckl and Holmes2007; Cottrell & Seckl, Reference Cottrell and Seckl2009; Wyrwoll & Holmes, Reference Wyrwoll and Holmes2012) and limited emerging human studies (Oberlander et al. Reference Oberlander, Weinberg, Papsdorf, Grunau, Misri and Devlin2008; O'Donnell et al. Reference O'Donnell, Bugge Jensen, Freeman, Khalife, O'Connor and Glover2012; Conradt et al. Reference Conradt, Lester, Appleton, Armstrong and Marsit2013; Moisiadis & Matthews, Reference Moisiadis and Matthews2014; Räikkönen et al. Reference Räikkönen, O'Reilly, Pesonen, Kajantie, Villa, Laivuori, Hämäläinen, Seckl and Reynolds2014; Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015) suggest that fetal overexposure to glucocorticoids may be a key underlying mechanism.
The placenta, which plays a vital role in mediating the maternal hormonal signals to the fetus, provides a barrier to fetal exposure to the much higher glucocorticoid levels in the maternal circulation (Seckl, Reference Seckl1998; Seckl & Meaney, Reference Seckl and Meaney2004; Seckl & Holmes, Reference Seckl and Holmes2007). This is ensured by the placental enzyme 11beta-hydroxysteroid dehydrogenase type 2 (11β-HSD2) which catalyzes metabolism of up to 80–90% of maternal active cortisol to inactive cortisone (Edwards et al. Reference Edwards, Benediktsson, Lindsay and Seckl1993). Among the other important placental regulators of feto-placental glucocorticoid overexposure are 11beta-hydroxysteroid dehydrogenase type 1 (11β-HSD1), which catalyzes regeneration of active cortisol from inactive cortisone, and intracellular glucocorticoid receptors (GRs) and mineralocorticoid receptors (MRs), which mediate glucocorticoid actions on gene transcription (Seckl, Reference Seckl1998; Seckl & Meaney, Reference Seckl and Meaney2004; Seckl & Holmes, Reference Seckl and Holmes2007; Cottrell & Seckl, Reference Cottrell and Seckl2009). Animal studies have shown that increased feto-placental exposure to active glucocorticoids of maternal origins, alters offspring brain anatomy, functioning of the hypothalamic-pituitary-adrenal (HPA) axis, hinders learning and memory, and increases anxiety- and depression-like behaviors of the offspring (Welberg et al. Reference Welberg, Seckl and Holmes2000; Holmes et al. Reference Holmes, Abrahamsen, French, Paterson, Mullins and Seckl2006; Wyrwoll & Holmes, Reference Wyrwoll and Holmes2012).
Fetal glucocorticoid exposure may also exert effects upon the development and subsequent function of the serotonergic nervous system of the offspring (Wyrwoll & Holmes, Reference Wyrwoll and Holmes2012), a key system implicated in mood disorders (Ressler & Nemeroff, Reference Ressler and Nemeroff2000). Glucocorticoid and serotonergic systems are known to interact such that glucocorticoids regulate serotonin synthesis, transport, re-uptake and neuronal receptor expression, while serotonin controls GR and MR expression in the central nervous system (Wyrwoll & Holmes, Reference Wyrwoll and Holmes2012). Recent data in the mouse show that placental deficiency of 11β-HSD2 is associated with increased serotonin synthesis and impairment in breakdown of serotonin in the brain of the offspring (Wyrwoll & Holmes, Reference Wyrwoll and Holmes2012).
Glucocorticoid (Murphy et al. Reference Murphy, Smith, Giles and Clifton2006) and serotonin (Bonnin et al. Reference Bonnin, Goeden, Chen, Wilson, King, Shih, Blakely, Deneris and Levitt2011) systems are both expressed in the placenta where interaction might occur and be relevant to exposure of the offspring's developing central nervous system. Yet, we are not aware of previous studies that have tested whether alterations in placental expression of key genes determining placental glucocorticoid and serotonin function play a role in programming of offspring behavioral outcomes and mediate the influence of prenatal maternal environmental adversity on these behavioral outcomes. Therefore, we tested if variations in placental mRNAs encoding 11β-HSD2 (HSD11B2), 11β-HSD1 (HSD11B1), GR (NR3C1), MR (NR3C2) and the serotonin transporter (SLC6A4) were associated with differences in regulatory behaviors of the offspring at a mean age of 15.6 days after birth. In the tests of mediation we focused on placental mRNA levels of NR3C1 and NR3C2 and on maternal depressive symptoms during pregnancy as the prenatal maternal environmental adversity as we have recently shown in this sample that maternal depressive symptoms during pregnancy were significantly associated with higher placental mRNA levels of NR3C1 and NR3C2, but were not significantly associated with the levels of HSD11B2, HSD11B1 or SLC6A4 (Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015). Consequently, our new analyses reported here extends our previous study by testing if placental NR3C1 and NR3C2 mRNAs mediate the influence of maternal depressive symptoms during pregnancy on infant regulatory behaviors, providing the other criteria for mediation were also met (Hayes, Reference Hayes2009), i.e. that placental NR3C1 and NR3C2 mRNAs and maternal depressive symptoms during pregnancy were also associated with infant regulatory behaviors.
Method
Participants
The participants were 67 healthy pregnant women enrolled in the Prediction and Prevention of Preeclampsia (PREDO) Study as described previously (Villa et al. Reference Villa, Kajantie, Räikkönen, Pesonen, Hämäläinen, Vainio, Taipale and Laivuori2013; Räikkönen et al. Reference Räikkönen, O'Reilly, Pesonen, Kajantie, Villa, Laivuori, Hämäläinen, Seckl and Reynolds2014; Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015). The women did not report using glucocorticoid or antidepressant medication during pregnancy, had no obstetric complications and delivered term (37 + 0–41 + 6 weeks of gestation), singleton, healthy infants. Of these, 54 participated in a follow-up study at a mean infant age of 15.6 [standard deviation (s.d.) = 4.3] days. Those who did not participate were similar to those who did (p > 0.11) except for a higher pre-pregnancy body mass index (BMI) [mean (s.d.) 27.1 (7.8) v. 22.9 (3.5) kg/m2, p = 0.004], placental weight [640.2 (102.2) v. 573.8 (83.2) g, p = 0.02] and shorter length of gestation [275.9 (6.5) v. 281.2 (8.6) days, p = 0.04] than participants. The study protocol was approved by the ethical committees at the Helsinki and Uusimaa Hospital District and written informed consent was obtained.
Maternal depressive symptoms
The Center for Epidemiologic Studies Depression Scale (CES-D; Radloff, Reference Radloff1977) for depressive symptoms was completed by the mothers bi-weekly during pregnancy from 12–13 gestational weeks onwards up to 14 times in total. The value at week 12 and mean values across weeks 14–26 and 28–38 were used as trimester-specific indices of depressive symptoms during pregnancy (Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015).
Placental tissue sampling and gene expression
Two sets of nine-site biopsies were collected from the decidual side of the placenta a maximum of 90 min after vaginal or cesarean delivery using standard protocols as previously described (Räikkönen et al. Reference Räikkönen, O'Reilly, Pesonen, Kajantie, Villa, Laivuori, Hämäläinen, Seckl and Reynolds2014; Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015). The biopsies were put in RNA-later (Qiagen GmbH, Germany) and stored at −20 °C. Total RNA was extracted from placental tissue sampled from a central site using Qiagen RNeasy mini kits (Qiagen Ltd, UK). The RNA concentration and purity of all samples was assessed using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, UK) and the integrity of RNA confirmed by separating rRNA using electrophoresis in a 1% agarose/0.5xTBE (45 mm Tris-borate, 1 mm EDTA) gel with 0.1 μl/ml Gel Red (Biotium, USA) at ~90 V for 30–50 min. cDNA synthesis was carried out using the Access RT–PCR system (Promega, UK). cDNA was incubated in triplicate with gene-specific primers and fluorescent probes either using the Universal Probe Library system from Roche Diagnostics Ltd (UK) for HSD11B1 (forward primer: caatggaagcattgttgtcg, reverse primer: ggcagcaaccattggataag) and NR3C2 (forward primer: tgggaattctgacttacttaacca, reverse primer: aatacaaaaagctgatgcagacc) or pre-designed assays from Applied Biosystems (ABI, USA) for NR3C1 (Hs00230818_m1), HSD11B2 (Hs00388669_m1) and SLC6A4 (Hs00984349_m1) in Roche Light Cycler 480 Probes mastermix. PCR cycling and detection of fluorescent signal was carried out using a Roche Light Cycler 480. A standard curve was constructed for each primer-probe set using a serial dilution of cDNA pooled from all samples. Results were corrected to the control gene TATA-binding protein (TBP).
Infant regulatory behaviors
Mothers rated infant regulatory behaviors using the Neonatal Perception Inventory (Broussard & Hartner, Reference Broussard, Hartner and Hellmuth1971). This inventory captures behaviors relating to infants’ crying, feeding, spitting, elimination (bowel movements), sleeping and predictability. In order to diminish the potential bias that may result in too positive or negative perceptions of infant behaviors, the mother was first asked to rate concerns relating to regulatory behaviors she would expect an ‘average’ infant to display. She was then asked to rate concerns in relation to her own infant's regulatory behaviors. The ratings were made using a five-point scale ranging from no problems to a great number of problems. A difference score between her own and the average infant's regulatory behaviors (own infant – average infant) reflects more regulatory challenges in her own infant's behavior. A principal components analysis revealed one factor (eigenvalue criterion >1) explaining 40% of the total variance, which lends credence to unidimensional structure of the scale and gives support to construct validity. Cronbach's alpha for internal consistency was 0.71 which gives support for the scale's reliability.
Covariates and confounders
Sample time between placental birth and biopsy (min) was recorded. Mode of delivery (vaginal v. elective/unplanned cesarean), parity (primiparous v. multiparous), maternal pre-pregnancy BMI (kg/m2), maternal age at delivery (years), smoking status during pregnancy (yes/no; number of cigarettes per day), infant sex (boy v. girl), gestation length (days) and infant birth weight (g) were derived from hospital birth records and the Finnish National Birth Register. Alcohol consumption during pregnancy (number of drinks during the past 4 weeks) and maternal level of education (primary, secondary, tertiary) were self-reported. The CES-D (Radloff, Reference Radloff1977) for depressive symptoms was completed by the mothers at the infant's age of 15.6 days.
Statistical analyses
Data were analyzed using IBM SPSS Statistics 22 for Windows (IBM Corp., USA). We first used linear regression analyses in testing associations between HSD11B2, HSD11B1, NR3C1, NR3C2, SLC6A4 mRNAs in term placenta and infant regulatory behaviors at age 15.6 days. We then tested if maternal depressive symptoms during pregnancy acted via altering glucocorticoid action in and via the placenta to impact on offspring regulatory behaviors by using the Mediate macro for SPSS with 5000 bootstrapped samples (Hayes & Preacher, Reference Hayes and Preacher2014). For these analyses gene expression data were log transformed to account for non-normality (after transformation skewness index divided by standard error <1.66 for all transformed variables) and thereafter the predictor and outcome variables were standardized to the mean of 0 and s.d. of 1 to facilitate interpretation. Hence, unstandardized regression coefficients and 95% confidence intervals (CIs) or test statistics represent effect sizes, s.d. unit per s.d. unit. We present the findings as unadjusted effect sizes and effect sizes from linear regression analyses when adjusted simultaneously for sampling time between placental birth and biopsy, mode of delivery, parity, maternal pre-pregnancy BMI, maternal age at delivery, level of education, smoking status and alcohol consumption during pregnancy, gestation length, infant birth weight and sex (model 1). In linear regression analyses we also made adjustments for maternal depressive symptoms measured at the infant's age of 15.6 days, as maternal depressive symptoms may bias perceptions (model 2).
Results
Table 1 shows clinical characteristics of the sample. Table 2 shows associations between covariates and confounders and infant regulatory behaviors. Mothers with a lower level of education and those who reported higher depressive symptoms in the third trimester and at infant age 15.6 days reported more infant regulatory behavioral challenges.
Table 1. Characteristics of the sample (N = 54)

CES-D, Center for Epidemiologic Studies Depression Scale.
a mRNA values are median (interquartile range).
b HSD11B2 refers to 11beta-hydroxysteroid dehydrogenase type 2.
c HSD11B1 refers to 11beta-hydroxysteroid dehydrogenase type 1.
d NR3C1 refers to glucocorticoid receptor.
e NR3C2 refers to mineralocorticoid receptor:
f SLC6A4 refers to serotonin transporter.
g <10 cigarettes per day, n = 1; <10 cigarettes per week or less often, n = 2.
h One drink refers to 33 cl beer or cider (3.5–4.7% vol alcohol), 12 cl mild wine (7–16% vol alcohol), 8 cl strong wine (15–22% vol), 4 cl liquor (>22% vol).
i Higher values indicate more regulatory behavioral challenges.
Table 2. Association between covariates and confounders and infant regulatory behaviors
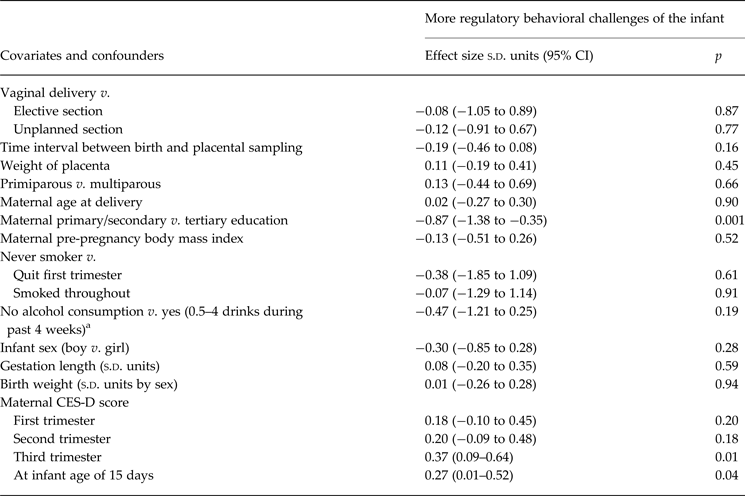
s.d. units, Standard deviation units; CI, confidence interval; CES-D, Center for Epidemiologic Studies Depression Scale.
a One drink of alcohol refers to 33 cl beer or cider (3.5–4.7% vol alcohol), 12 cl mild wine (7–16% vol alcohol), 8 cl strong wine (15–22% vol), 4 cl liquor (>22% vol).
Placental mRNA levels and infant regulatory behaviors
Table 3 shows that higher placental mRNA levels of HSD11B2, HSD11B1, NR3C1, SLC6A4, but not NR3C2, were significantly associated with more regulatory behavioral challenges of the infant: the increase was 0.41, 0.30, 0.44 and 0.26 s.d. units per each s.d. unit increase in the respective mRNA level (p values < 0.05). Fig. 1 displays the regression lines and 95% CIs of the significant unadjusted associations and additionally shows that the proportion of variance in infant regulatory behaviors accounted for by placental HSD11B2, HSD11B1, NR3C1 and SLC6A4 mRNAs varied from 8.9% to 19.9%.
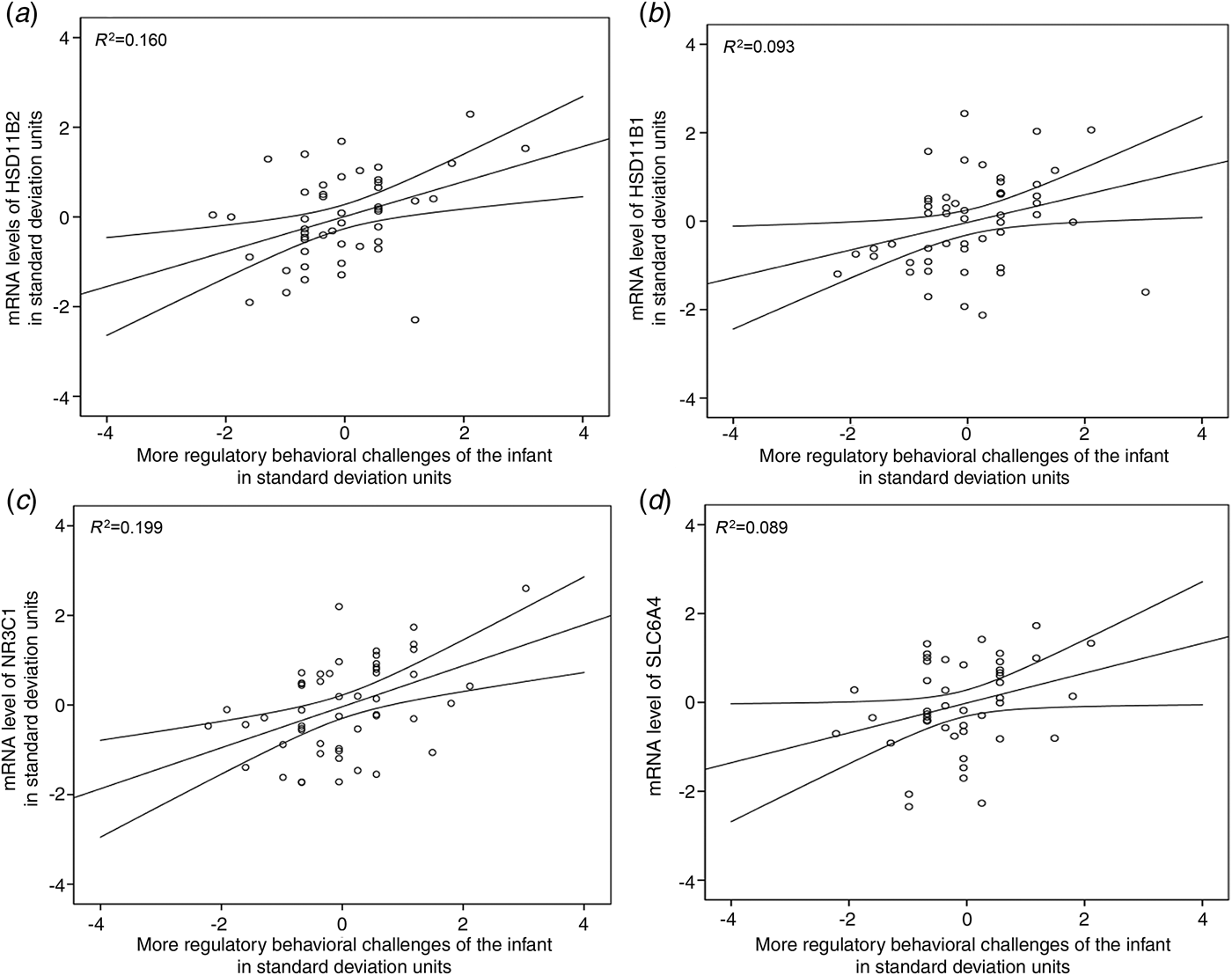
Fig. 1. Unadjusted associations between mRNA levels of (a) 11beta-hydroxysteroid dehydrogenase type 2 (HSD11B2), (b) 11beta-hydroxysteroid dehydrogenase type 1 (HSD11B1), (c) glucocorticoid receptor (NR3C1) and (d) serotonin transporter (SLC6A4) in term placenta and infant regulatory behavioral challenges. The lines represent unadjusted unstandardized regression coefficients and 95% confidence intervals; R 2 refers to the proportion of variance in infant regulatory behaviors accounted for by the placental mRNA levels.
Table 3. Associations between mRNA levels of 11beta-hydroxysteroid dehydrogenase type 2 (HSD11B2), 11beta-hydroxysteroid dehydrogenase type 1 (HSD11B1), glucocorticoid receptor (NR3C1), mineralocorticoid receptor (NR3C2) and serotonin transporter (SLC6A4) in term placenta with infant regulatory behaviors

s.d. units, Standard deviation units; CI, confidence interval.
a Model 1 refers to adjustments made for mode of delivery, time difference between placental birth and biopsy, maternal age at delivery, maternal education, parity, maternal pre-pregnancy body mass index, smoking and alcohol consumption during pregnancy, gestation length, birth weight by sex and infant sex.
b Model 2 refers to additional adjustments made for maternal depressive symptoms (Center of Epidemiological Studies Depression Scale score) at infant age of 15.6 days.
Table 3 shows that when we made adjustments for sampling time between placental birth and biopsy, mode of delivery, parity, maternal pre-pregnancy BMI, maternal age at delivery, level of education, smoking status and alcohol consumption during pregnancy, gestation length, infant birth weight and sex (model 1) the association with HSD11B1 was rendered non-significant; When the associations were adjusted for maternal depressive symptoms at infant age 15.6 days (model 2) the association with SLC6A4 was rendered non-significant (Table 3).
Mediation analyses
As placental NR3C2 mRNA level was not significantly associated with infant regulatory behaviors (Table 3) mediation via NR3C2 mRNA expression was not tested. Similarly, since maternal first and second trimester depressive symptoms were not significantly associated with infant regulatory behaviors (Table 2) they were not included in the mediation analyses. As maternal depressive symptoms during the third pregnancy trimester, placental NR3C1 mRNA level, and infant regulatory behaviors were all significantly inter-related (Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015) (Tables 2 and 3), and hence the criteria for testing mediation were met (Hayes, Reference Hayes2009; Hayes & Preacher, Reference Hayes and Preacher2014), we pursued in testing if NR3C1 mRNA mediated the effect of maternal depressive symptoms during the third pregnancy trimester on infant regulatory behaviors. Indeed, the indirect path via placental NR3C1 mRNA was statistically significant (indirect effect = 0.15, 95% CI 0.02–0.38, p < 0.05) (Fig. 2). Yet, the association between maternal depressive symptoms and infant regulatory behaviors was not rendered to zero, but remained ‘marginally’ significant (p = 0.08) (Fig. 2). This suggested that placental NR3C1 mRNA partly mediated the effect of maternal depressive symptoms on infant regulatory behaviors (Fig. 2). Fig. 2 also shows that the proportion of variance in NR3C1 mRNA accounted for by maternal depressive symptoms was 14.4% (p = 0.006) and the proportion of variance in infant regulatory behaviors accounted for by maternal depressive symptoms and placental NR3C1 mRNA was 24.8% (p = 0.0009). Adjustment for covariates and confounders did not alter the significant paths (p values < 0.05) or mediation (indirect effect = 0.13, 95% CI 0.01–0.34, p < 0.05 in model 1; indirect effect = 0.10, 95% CI 0.01–0.30, p < 0.05 in model 2).
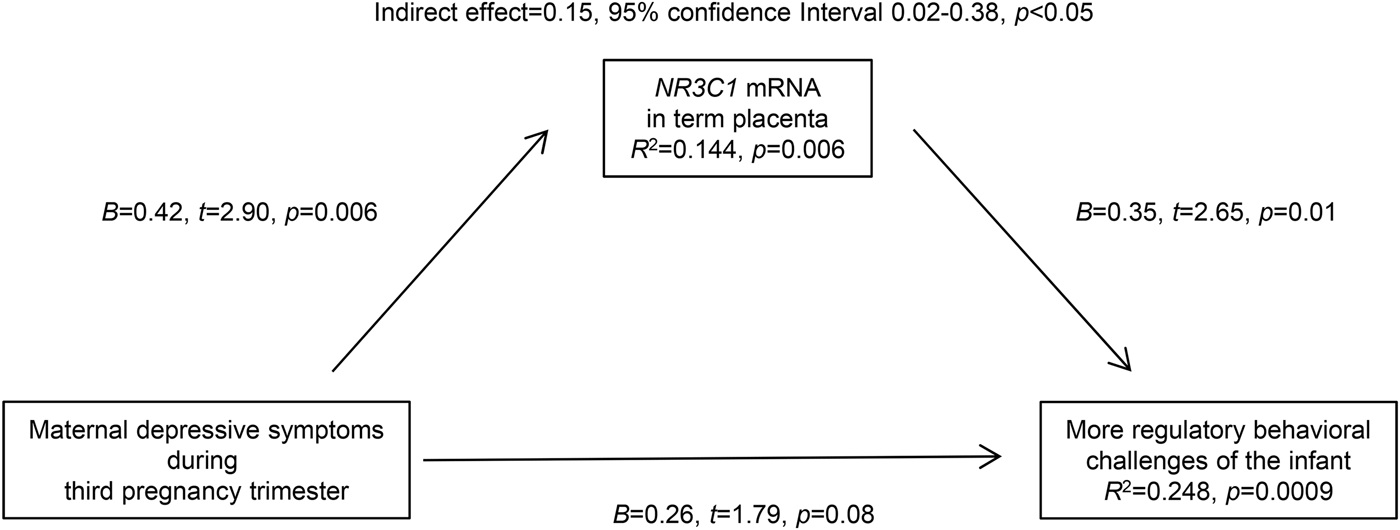
Fig. 2. Mediation analyses results showing that maternal depressive symptoms during the third trimester of pregnancy partly act via altering expression of glucocorticoid receptor (NR3C1) mRNA levels in term placenta to impact on infant regulatory behaviors. Numbers represent unadjusted unstandardized coefficients, 95% confidence intervals, and p values; R 2 refers to the proportion of variance in infant regulatory behaviors accounted for by the placental mRNA levels of NR3C1 and maternal depressive symptoms, and in placental mRNA levels of NR3C1 by maternal depressive symptoms.
Discussion
We show that variation in the mRNA levels of key gene products determining glucocorticoid and serotonin function in term human placenta is associated with more regulatory behavioral challenges of the infant at 15.6 days. More specifically, higher placental mRNA levels of HSD11B2, HSD11B1, NR3C1 and SLC6A4 were associated with infants who subsequently showed greater behavioral challenges in crying, sleeping, feeding, spitting and/or elimination. The findings with NR3C1 and HSD11B1 add increased placental sensitivity to glucocorticoids coupled with local amplification of glucocorticoid action within the placenta as a mechanism linking increased glucocorticoid exposure to adverse offspring outcomes. Yet, the finding of higher placental mRNA level of HSD11B2 was contrary to what we expected based on evidence from animal studies where fetal glucocorticoid overexposure has been secondary to down-regulation of placental HSD11B2 gene expression and activity, and hence inhibition of the function of the placental glucocorticoid barrier (Edwards et al. Reference Edwards, Benediktsson, Lindsay and Seckl1993; Lindsay et al. Reference Lindsay, Lindsay, Waddell and Seckl1996a , Reference Lindsay, Lindsay, Edwards and Seckl b ; Benediktsson et al. Reference Benediktsson, Calder, Edwards and Seckl1997; Seckl, Reference Seckl1998; Welberg et al. Reference Welberg, Seckl and Holmes2000; Seckl & Meaney, Reference Seckl and Meaney2004; Holmes et al. Reference Holmes, Abrahamsen, French, Paterson, Mullins and Seckl2006; Seckl & Holmes, Reference Seckl and Holmes2007; Wyrwoll & Holmes, Reference Wyrwoll and Holmes2012). As HSD11B2 excludes glucocorticoids from the syncytiotrophoblast we can only speculate that our finding showing that the placental mRNA level of HSD11B2 was higher in infants with more behavioral challenges may be an adaptive placental response to ameliorate the higher glucocorticoid availability and sensitivity in the placental compartment. Moreover the associations with NR3C1 and HSD11B2 remained significant when we made adjustments for several important factors that we have previously shown to relate to placental gene expression (Räikkönen et al. Reference Räikkönen, O'Reilly, Pesonen, Kajantie, Villa, Laivuori, Hämäläinen, Seckl and Reynolds2014; Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Laivuori, Villa, Hämäläinen, Seckl and Räikkönen2015) and/or infant regulatory behaviors, including mode of delivery, time difference between placental birth and biopsy, maternal age at delivery, parity, maternal pre-pregnancy BMI, smoking and alcohol consumption during pregnancy, maternal level of education, gestation length, infant birth weight and sex and maternal depressive symptoms measured in conjunction with infant behavioral ratings. In the models including maternal depressive symptoms at the time of making the infant ratings, HSD11B1 remained significant while SLC6A4 did not, and in the models including the other covariates SLC6A4 remained significant while HSD11B1 did not.
Our study is also the first to show that higher placental NRC31 mRNA level plays a role in the process that may partly mediate the effects of higher maternal depressive symptoms during the third trimester of pregnancy on more regulatory behavioral challenges of the infant. Statistical mediation via NR3C1 mRNA partially suggests that other factors may also be involved. These factors may include other gene products, such as the corticotropin-releasing hormone, or epigenetic modifications involved in the signaling and interactions of glucocorticoids and serotonin between the mother, placenta and fetus. Indeed, a recent study demonstrated that higher maternal self-reported depression during pregnancy and higher placental methylation of the NR3C1 were associated with poorer self-regulation, more hypotonia and more lethargy in the infant (Conradt et al. Reference Conradt, Lester, Appleton, Armstrong and Marsit2013). Yet, it remains unknown what the environmental exposures are that underlie the higher placental mRNA levels of HSD11B1, HSD11B2 and SLC6A4. While in this study maternal depressive symptoms during pregnancy were not associated with mRNA levels of these genes and therefore we did not pursue testing them as mediators, one previous study has shown that higher maternal anxious but not depressed mood a day before an elective cesarean section was associated with lower placental HSD11B2 mRNA level (O'Donnell et al. Reference O'Donnell, Bugge Jensen, Freeman, Khalife, O'Connor and Glover2012), and one other study has demonstrated that a history of maternal depressed mood during pregnancy, irrespective of the maternal antidepressant medication use, was associated with higher placental SLC6A4 mRNA level, but not with HSD11B2 (Ponder et al. Reference Ponder, Salisbury, McGonnigal, Laliberte, Lester and Padbury2011). Further studies are clearly warranted that unravel the maternal-placental-fetal pathways underpinning individual differences in early infant behaviors.
While we studied healthy term pregnancies, term placentas and healthy infants, our findings may have clinical relevance. Infant regulatory behaviors are among the earliest signs of neurobehavioral and neuropsychiatric problems in later life. Extensive data, including studies using the Neonatal Perception Inventory (the measure as used in our study), suggest that more regulatory behavioral challenges of the infant as reported by the mother are associated with later behavioral problems, including poorer cognitive functioning (Wolke et al. Reference Wolke, Schmid, Schreier and Meyer2009) and increased risk of externalizing behavior problems (Wolke et al. Reference Wolke, Rizzo and Woods2002) and attention deficit hyperactivity disorder (Wolke et al. Reference Wolke, Rizzo and Woods2002; Hemmi et al. Reference Hemmi, Wolke and Schneider2011) in childhood. Extension of our findings into the prevalence of neurobehavioral and neuropsychiatric problems in childhood in this sample is subject to ongoing studies.
We used mothers’ reports of infant regulatory behavioral challenges. This may introduce a bias that results in too positive or negative perceptions of infant behaviors. To overcome any potential bias in reporting, the mothers were asked to rate regulatory behaviors they would expect an ‘average’ infant to display and then rate her own infant. In addition, we made adjustments for maternal depressive symptoms at the time of infant's assessment and the mother's level of education that were expected to be associated with greater behavioral challenges of the infant. The majority of previous studies have, however, used mother- or parent-reports of infant regulatory behavioral challenges, as we did. Neurologists’ assessments have been used in some studies, but these assessments usually concern oral motor functioning in relation to feeding problems (Schmid et al. Reference Schmid, Schreier, Meyer and Wolke2010). Hence, use of mother-reports remains a limitation, as long as gold standards do not exist on how to measure regulatory behavioral challenges in infancy.
We recognize that we lack data on several elements of the maternal-placental-fetal glucocorticoid and serotonin signaling pathway. As we lack data on fetal and intra-placental glucocorticoid and serotonin levels, we cannot determine the degree of maternal glucocorticoids and serotonin transmitted via the placenta to the fetus. However, while maternal and fetal cortisol levels are highly correlated, a recent study has demonstrated that during prenatal life, most fetal serotonin is of placental and fetal, not of maternal, origins (Bonnin et al. Reference Bonnin, Goeden, Chen, Wilson, King, Shih, Blakely, Deneris and Levitt2011). We also have no information of any cell-specificity of actions and changes in the placenta itself, although the full thickness placental biopsies will principally reflect fetal characteristics. We only measured mRNA levels of the key genes of interest and lack data on levels of their respective protein levels. Further studies are needed to determine the mechanisms of altered gene expression, such as methylation of promoter and transcription factor-binding sites and other epigenetic mechanisms, and whether these are modifiable, as are studies that focus on functional end products of the key genes of interest. Other study limitations relate to the external validity of the findings. As the studied sample comprised healthy term pregnancies, healthy babies and very few mothers reported depressive symptoms that were above the clinical cut-off of the CES-D scale, our findings may not generalize to samples with greater variation in adversity.
In conclusion, our findings suggest that higher placental expression of key genes determining glucocorticoid and serotonin function in term human placenta and regulating feto-placental glucocorticoid and serotonin exposure is characteristic of infants with more regulatory behavioral challenges, particularly challenges relating to crying, sleeping, feeding, spitting and elimination behaviors. Our findings also suggest that higher placental mRNA levels of NR3C1 play a role in the process that may partly mediate the effects of maternal depressive symptoms during the third trimester of pregnancy on infant regulatory behavioral challenges. Our findings add placental glucocorticoid and serotonergic function as novel processes that may underpin developmental programming of early infant behaviors.
Acknowledgements
The study was supported by the Academy of Finland, the Signe and Ane Gyllenberg Foundation, the Emil Aaltonen Foundation, EVO (a special state subsidy for health science research), the Finnish Medical Foundation, the Jane and Aatos Erkko Foundation, the Novo Nordisk Foundation, the Päivikki and Sakari Sohlberg Foundation the Sigrid Juselius Foundation, and the Sir Jules Thorn Charitable Trust, University of Helsinki. We gratefully acknowledge the expertise of Dr Niina Komsi and Research Nurse Miia Tommola for data collection.
Declaration of Interest
None.







