Introduction
The family Arthoniaceae has a worldwide distribution and includes lichenized, lichenicolous and saprobic species (e.g. Tehler Reference Tehler1990; Sundin & Tehler Reference Sundin and Tehler1998; Sundin Reference Sundin1999; Grube Reference Grube2001, Reference Grube, Nash, Gries and Bungartz2007; Frisch et al. Reference Frisch, Thor, Ertz and Grube2014a). It is the largest family in the order Arthoniales and the seventh largest family in terms of number of lichenized species (Lücking et al. Reference Lücking, Hodkinson and Leavitt2017). The family is undergoing major changes following molecular analyses, with large genera being split into smaller entities (e.g. Frisch et al. Reference Frisch, Thor, Ertz and Grube2014a, Reference Frisch, Ohmura, Ertz and Thor2015; Van den Broeck & Ertz Reference Van den Broeck and Ertz2016; Van den Broeck et al. Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018).
In 2011, a lichenological field meeting was organized in Luxembourg as part of the annual field meeting of the association ‘Werkgroep Bryologie en Lichenologie’ of Flanders (Belgium). A total of nine localities were visited in the Ardenne district (Oesling), a massif of markedly siliceous rocks dating back to the Cambrian, Ordovician and Lower Devonian, covered by extensive forests. In the course of three days, a total of 253 lichens and lichenicolous fungi were recorded (Van den Broeck et al. Reference Van den Broeck, Diederich and Ertz2013). A striking species having a leproid thallus with a trentepohlioid photobiont was collected on siliceous rocks and could not be identified. The localities were revisited in 2018 to collect additional fresh specimens for sequencing, in order to determine its generic position. The present study aims to describe this new species in the genus Synarthonia with the support of molecular data. An additional sequence obtained from British material of Arthonia atlantica P. James, which belongs to the same subclade of Arthoniaceae as the new species, was added to the phylogenetic analyses to determine its generic affiliation. The phylogenetic positions of Reichlingia anombrophila (Coppins & P. James) Frisch and Synarthonia astroidestera (Nyl.) Ertz & Van den Broeck are shown for the first time.
Materials and Methods
Voucher specimens are deposited in the herbaria BR and E, and in the private collections of N. Sanderson and P. Diederich. The external morphology was studied and measured using an Olympus SZX12 stereomicroscope. Macroscopic images were captured with a Keyence VHX-5000 digital microscope and a VH-Z20R/W/T lens. Hand-cut sections and squash preparations of thallus were mounted in water, 5% aqueous potassium hydroxide solution (K), or Lugol's iodine solution (1% I2) without (I) or with K pretreatment (KI) and observed using an Olympus BX51 compound microscope. Measurements refer to dimensions in K. Microscopic images were captured using an Olympus BX51 compound microscope fitted with an Olympus SC50 digital camera. Colour reactions of the thallus were studied using K, common household bleach (C), K followed by common household bleach (KC), crystals of para-phenylenediamine dissolved in ethanol (PD) and longwave UV (366 nm). Lichen secondary metabolites were identified using thin-layer chromatography (TLC) in solvents B, C and G (Orange et al. Reference Orange, James and White2010).
Molecular techniques
Well-preserved and freshly collected specimens lacking any visible symptoms of fungal infection were used for DNA isolation. Hand-cut sections of the apothecia or a small number of soredia were used for direct PCR as described in Ertz et al. (Reference Ertz, Tehler, Irestedt, Frisch, Thor and van den Boom2015). The lichen material was washed with acetone and then rinsed with water to remove remnants of pigments. The material was placed directly in microtubes with 20 μl H2O. Amplification reactions were prepared for a 50 μl final volume containing 5 μl 10× DreamTaqBuffer (Thermo Scientific, www.thermoscientific.com/onebio), 1.25 μl of each of the 20 μM primers, 5 μl of 2.5 mg ml−1 bovin serum albumin (Thermo Scientific), 4 μl of 2.5 mM dNTPs (Thermo Scientific), 1.25 U DreamTaq DNA polymerase (Thermo Scientific) and the lichen material. A targeted fragment of c. 0.8 kb of the mtSSU rDNA was amplified using primers mrSSU1 and mrSSU3R (Zoller et al. Reference Zoller, Scheidegger and Sperisen1999). A fragment of c. 1 kb of the RPB2 protein-coding gene was amplified using primers fRPB2-7cF and fRPB2-11aR (Liu et al. Reference Liu, Whelen and Hall1999). The yield of the PCR reactions was verified by running the products on a 1% agarose gel using ethidium bromide for visualization. Both strands were sequenced by Macrogen® using amplification primers. Sequence fragments were assembled with Sequencher v.5.4.6 (Gene Codes Corporation, Ann Arbor, Michigan). Sequences were subjected to ‘Megablast’ searches in GenBank to verify their closest relatives and to detect potential contaminations.
Taxon selection and phylogenetic analyses
Seven new mtSSU sequences from four species and four RPB2 sequences from three species were obtained for this study (Table 1). For the phylogenetic analyses, the sequences were included in the mtSSU and the RPB2 datasets published by Van den Broeck et al. (Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018), consisting of taxa representing all major clades currently accepted in the Arthoniaceae except for the more distantly related Bryostigma clade (Frisch et al. Reference Frisch, Thor, Ertz and Grube2014a). The sequences were aligned using MAFFT v.7.402 (Katoh et al. Reference Katoh, Misawa, Kuma and Miyata2002) on the CIPRES Web Portal (Miller et al. Reference Miller, Pfeiffer and Schwartz2010) and manually corrected for errors using Mesquite 3.04 (Maddison & Maddison Reference Maddison and Maddison2015). Arthothelium norvegicum Coppins & Tønsberg was chosen as outgroup species. Terminal ends of sequences and ambiguously aligned regions were delimited manually and excluded from the datasets.
Table 1. Species names, voucher specimens, countries of origin and GenBank Accession numbers, of sequences generated in this study, for the specimens included in the phylogenetic inference (Fig. 1). GenBank Accession numbers for the other taxa in Fig. 1 are listed in Van den Broeck et al. (Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018).
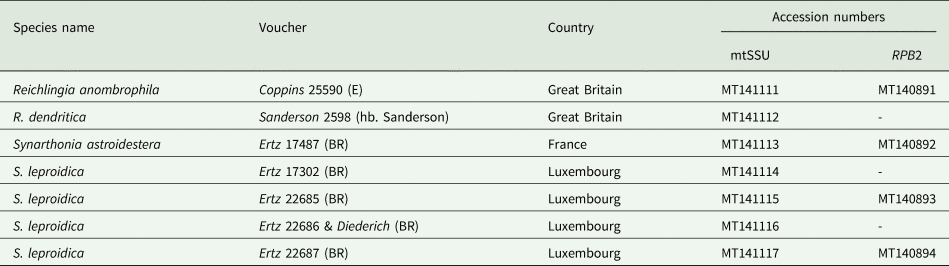

Fig. 1. Phylogenetic relationships among a selected group of Arthoniaceae resulting from a Bayesian analysis based on a dataset of 56 samples of mtSSU and RPB2 sequences. Arthothelium norvegicum was chosen as outgroup. MrBayes posterior probabilities are shown above branches, and RAxML bootstrap values are shown below branches. Thicker lines highlight internodes considered as strongly supported by both analyses. Names of samples for which sequences were generated in this study are indicated in bold. The new species of Synarthonia is highlighted with a shaded box. In colour online.
To examine topological incongruence among datasets, a maximum likelihood (ML) analysis was carried out on each of the single-locus datasets. We used RAxML v.8.2.12 (Stamatakis Reference Stamatakis2014) with 1000 replicates of ML bootstrapping (ML-BS) under the GTRGAMMA model of sequence evolution. Analyses were run on the CIPRES Web Portal (Miller et al. Reference Miller, Pfeiffer and Schwartz2010). All topological bipartitions were compared for the two loci. A conflict was assumed to be significant when differing topologies for the same set of taxa (one being monophyletic and the other being non-monophyletic) were each supported with bootstrap values ≥ 70 (Mason-Gamer & Kellogg Reference Mason-Gamer and Kellogg1996). Based on this criterion, the same conflict as already highlighted by Van den Broeck et al. (Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018) was detected regarding the sister-group relationship of Synarthonia with either Coniocarpon or Reichlingia. As this conflict had no impact on the monophyly of either Reichlingia or Synarthonia, and thus on our conclusions regarding the generic affiliations of the newly sequenced species, the mtSSU and RPB2 datasets were concatenated.
The combined two-locus dataset of 56 samples consisted of 1503 unambiguously aligned sites, 639 for mtSSU and 864 for RPB2. Bayesian analyses were carried out on the concatenated two-locus dataset using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method in MrBayes v.3.2.7a (Huelsenbeck & Ronquist Reference Huelsenbeck and Ronquist2001; Ronquist & Huelsenbeck Reference Ronquist and Huelsenbeck2003) on the CIPRES Web Portal (Miller et al. Reference Miller, Pfeiffer and Schwartz2010). Best-fit evolutionary models for each partition were estimated using the Akaike Information Criterion (AIC) as implemented in jModelTest2 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012). The GTR + I + G model was selected for the mtSSU dataset as well as for the RPB2/1st position, while the TVM + I + G model was selected for the RPB2/2nd and RPB2/3rd positions. Two Bayesian MCMCMC runs were executed in parallel, each using four independent chains and 120 million generations, sampling trees every 1000th generation. Tracer v.1.6.0 (Rambaut et al. Reference Rambaut, Suchard, Xie and Drummond2013) was used to ensure that convergence was reached by plotting the log-likelihood values of the sample points against generation time. Convergence between runs was also verified using the PSRF (Potential Scale Reduction Factor), confirming that values for all parameters were equal to 1.000. Posterior probabilities (PP) were determined by calculating a majority-rule consensus tree generated from the 180 002 post burn-in trees out of the 240 002 trees sampled by the two MCMCMC runs using the sumt option of MrBayes. In addition, a maximum likelihood (ML) analysis was performed using RAxML v.8.2.12 (Stamatakis Reference Stamatakis2014) with 1000 ML bootstrap iterations (ML-BS) and the GTRGAMMA model.
The Bayesian tree did not contradict the RAxML tree topology for the strongly supported branches. Therefore, only the Bayesian tree is shown, with the PP values added above the internal branches and the ML-BS values added below (Fig. 1). Internodes with ML-BS ≥ 70 and PP ≥ 95 were considered strongly supported (Alfaro et al. Reference Alfaro, Zoller and Lutzoni2003; Lutzoni et al. Reference Lutzoni, Kauff, Cox, McLaughlin, Celio, Dentinger, Padamsee, Hibbett, James and Baloch2004). Phylogenetic trees were visualized using FigTree v.1.4.2 (Rambaut Reference Rambaut2012).
Results and Discussion
Phylogenetic analyses
The Bayesian tree obtained from the combined two-locus analysis of 56 samples is shown in Fig. 1. The main well-supported lineages of Arthoniaceae are in accordance with the results obtained by Van den Broeck et al. (Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018). The genus Synarthonia is placed in a well-supported lineage (ML-BS = 96 and PP = 1) with the genus Coniocarpon and the Reichlingia group, both also being well-supported monophyletic groups (ML-BS = 98–100 and PP = 1) (Fig. 1). The new species is nested within the genus Synarthonia, as sister taxon to S. muriformis. It belongs to the core group of Synarthonia (i.e. from S. astroidestera to S. inconspicua (Stirt.) Van den Broeck & Ertz) characterized by species having notably white pruinose ascomata (as defined by Van den Broeck et al. Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018). This core group is well supported in our phylogenetic tree (ML-BS = 99 and PP = 1). Our new sequences further confirm the placement of Arthonia astroidestera in the genus Synarthonia. This species was transferred from Arthonia to Synarthonia by Van den Broeck et al. (Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018) based on morphology and chemistry alone. The newly sequenced Arthonia atlantica clusters with species of Reichlingia in a strongly supported lineage (ML-BS = 77 and PP = 1) and is thus transferred to this genus (see Taxonomy section). Arthonia anombrophila was recently transferred to the genus Reichlingia by Frisch et al. (Reference Frisch, Klepsland, Palice, Bendiksby, Tønsberg and Holien2020) based on unpublished sequences from a Norwegian specimen. Our new sequences from British material confirm its placement in Reichlingia. Relationships within the genus Reichlingia are poorly supported. The generic affiliation of A. anglica published by Ertz et al. (Reference Ertz, Miadlikowska, Lutzoni, Dessein, Raspé, Vigneron, Hofstetter and Diederich2009; sub ‘Arthonia sp. 1’) is in need of further studies with more data, and the identity of the sequenced specimen requires confirmation because the sample comes from tropical Africa while the type locality of A. anglica is in Great Britain.
Taxonomy
Synarthonia leproidica Ertz, Aptroot & Diederich sp. nov.
MycoBank No.: MB 834820
A species of Synarthonia characterized within the genus by a pale greyish leproid thallus with a dark brown violaceous tinge at the surface, psoromic acid as a thallus compound, a saxicolous habit and a phylogenetic position sister to Synarthonia muriformis.
Type: Luxembourg, Lellingen, Vallée du Lellgerbaach, Op Bärel, 49°59′11″N, 6°01′16″E, 323 m elev., chênaie-charmaie de bas de pente exposée nord-nord-ouest, petite paroi siliceuse éclairée au bord d'un chemin forestier, sur face rocheuse subverticale plus ou moins abritée de la pluie, 8 September 2018, Ertz 22686 & Diederich (BR—holotype!; hb. Diederich—isotype!).

Fig. 2. Synarthonia leproidica (B & E, Ertz 22687; C & F, Ertz 22686 – holotype; D, Diederich 18666). A, habitat in Lellingen (Luxembourg). Type locality where S. leproidica is abundant on the rocks. B–D, thallus. E & F, soredia in K. Scales: B = 2.5 mm; C = 0.8 mm; D = 1 mm; E & F = 10 μm. In colour online.
Thallus saxicolous, crustose, leproid, forming patches of c. 0.5–5 mm diam., or confluent and often covering large areas up to c. 10 cm diam., up to 1(–2) mm thick, pale greyish, with dark brown violaceous tinge at the surface, and a greenish tinge inside when abraded (in thicker parts of the thallus). Soredia (14–)20–38 μm diam., formed of individual or short chains of photobiont cells surrounded by hyaline to dark brown hyphae of 2(–3) μm diam.; soredia without projecting hyphae, with many hyaline crystals of c. 0.5–1.5 μm diam. visible on the hyphae in polarized light that dissolve in K. Photobiont trentepohlioid, containing orange pigments, visible as individual globose cells, (6–)9–15(–25) μm diam. or in short chains of c. 2–4 cells, with individual elliptical to rectangular algal cells of 13–20(–29) × (5–)9–16 μm.
Ascomata and conidiomata unknown.
Chemistry
Thallus K−, C−, PD+ bright orange-yellow, UV−; hyphae I+ pale orange, KI+ pale orange. TLC revealed the presence of psoromic acid (specimen Ertz 17302 tested in solvents B and G; Ertz 22685, 22686, 22687 tested in solvent C).
Etymology
The specific epithet refers to the leproid thallus.
Distribution and ecology
So far known only from three localities in Luxembourg, where it inhabits sheltered siliceous schistose rock faces in rather open conditions near forest edges. The associated lichen vegetation is species poor and also includes Psilolechia lucida (Ach.) M. Choisy and Lepraria spp.
Discussion
Synarthonia leproidica is a saxicolous sterile leproid crustose lichen with a trentepohlioid photobiont and a greyish thallus with a dark brown violaceous tinge reacting PD+ bright orange-yellow. Our molecular data surprisingly place the species within the genus Synarthonia (Fig. 1). The new species is at present the only known leproid member of the genus and also the only saxicolous Synarthonia (Van den Broeck et al. Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018). Synarthonia sikkimensis S. Joseph & G. P. Sinha is the only known species of the genus having a sorediate thallus. In that species, the thallus is never leproid, but discrete small soralia are present near the margins of the thallus. Moreover, S. sikkimensis differs from the new species by a well-developed rhizomorph-like prothallus and the absence of secondary metabolites in the thallus (Joseph & Sinha Reference Joseph and Sinha2015). Psoromic acid is a substance present in the new species but also in several other species of Synarthonia (e.g. S. borbonica (Ertz et al.) Van den Broeck & Ertz, S. psoromica S. Joseph & G. P. Sinha, S. muriformis Van den Broeck et al.). In our phylogenetic analyses, it is notable that the closest relative of S. leproidica is S. muriformis which shares psoromic acid but its chemistry is more varied with the presence of evernic acid and two unknown UV+ white unidentified secondary compounds (Van den Broeck et al. Reference Van den Broeck, Frisch, Razafindrahaja, Van de Vijver and Ertz2018).
Siliceous rock outcrops in the Luxembourg part of the Ardennes are surprisingly rich in sterile Arthoniales. Dendrographa latebrarum (Ach.) Ertz & Tehler and Sparria endlicheri (Garov.) Ertz & Tehler are other strictly sterile Arthoniales while others are lichenized hyphomycetes such as Milospium deslooveri Diederich & Sérus. and Reichlingia leopoldii Diederich & Scheid. The discovery of the new species suggests that other strictly sorediate lichen species might have been overlooked on siliceous rocks in European regions, even in intensely explored countries such as Luxembourg.
Snippocia nivea (D. Hawksw. & P. James) Ertz & Sanderson is another strictly sterile member of the Arthoniaceae known from Western Europe containing psoromic acid but it differs from the new species by a corticolous pale pink thallus (James Reference James1971) and is unrelated to the genus Synarthonia in molecular studies (Ertz et al. Reference Ertz, Sanderson, Łubek and Kukwa2018). Roccellographa sorediata (Sparrius et al.) Coppins & Fryday is a common saxicolous, usually sterile, ±entirely sorediate lichen also producing psoromic acid, but it differs from the new species by a milk white to grey thallus with a conspicuous black prothallus and inhabits coastal rock faces (Fletcher Reference Fletcher, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009).
Additional specimens examined
Luxembourg: Lellingen, Vallée du Lellgerbaach, Op Bärel, 49°59′25″N, 6°02′03″E, 350 m elev., 2011, Ertz 17302 (BR) & Diederich 17236 (hb. Diederich); ibid., 2018, Ertz 22685 (BR) & Diederich 18665 (hb. Diederich); same locality as type, 2018, Diederich 18666 (hb. Diederich); Bourscheid, sentier autour du château, 49°54′21″N, 6°04′49″E, 362 m elev., petite paroi de rocher siliceux ombragée sous le mur d'enceinte du château, sur face rocheuse subverticale ± abritée de la pluie, 2018, Ertz 22687 (BR) & Diederich 18663 (hb. Diederich).
Reichlingia
The genus Reichlingia was originally monotypic and described as a lichenicolous fungus (Diederich & Scheidegger Reference Diederich and Scheidegger1996) but is now generally considered to be a lichenized hyphomycete (Diederich & Coppins Reference Diederich, Coppins, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). The type species has a byssoid thallus and produces dark brown, multicellular, branched conidia grouped in irregular sporodochia-like conidiomata on its upper surface (Diederich & Coppins Reference Diederich, Coppins, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). The genus has been emended to include three fertile species of Arthoniaceae (Frisch et al. Reference Frisch, Thor and Sheil2014b) and Arthonia anombrophila was recently transferred to Reichlingia (Frisch et al. Reference Frisch, Klepsland, Palice, Bendiksby, Tønsberg and Holien2020). Here, DNA sequence data support A. atlantica as an additional species in this genus and the phylogenetic position of R. anombrophila is shown for the first time (see ‘Phylogenetic analyses’ above). The morphology of both species fits well with the current concept of the genus Reichlingia, although R. anombrophila differs from the other species by having an immersed or partly superficial, compact thallus.
Reichlingia dendritica (Leight.) Ertz & Sanderson comb. nov.
MycoBank No.: MB 834821
Stigmatidium dendriticum Leight., in J. Bot., Lond. 13, 257 (1875).—Arthonia dendritica (Leight.) Cromb., J. Bot., Lond. 14, 362 (1876), non (Ach.) Duf. (1818).—Enterographa dendritica (Leight.) P. James, Lichenologist 3, 97 (1965).—Arthonia atlantica P. James, Lichenologist 4, 318 (1970); type: not seen.
Notes
Arthonia dendritica (Leight.) Cromb. was illegitimate because of the earlier homonym Arthonia dendritica (Ach.) Duf. (=Phaeographis dendritica (Ach.) Müll. Arg.). Therefore, James (Reference James1970) published the replacement name Arthonia atlantica P. James for accepting the species in Arthonia. Following our phylogenetic results where A. atlantica is the sister taxon of Reichlingia syncesioides Frisch & G. Thor (Fig. 1), a new combination is made in Reichlingia using the epithet from the basionym (Stigmatidium dendriticum Leight.), while Arthonia atlantica becomes one of its homotypic synonyms.
Sequenced specimen
Great Britain: Wales: V.C.48, Merionethshire, Coed Maentwrog NNR, Coed Glan-yr-afon, Grid Ref. SH67465 41609, 70 m elev., rock face in oceanic pasture woodland, overhanging slate outcrop, 2019, Sanderson 2598 (hb. Sanderson).
Acknowledgements
We wish to thank very warmly Myriam Dehaan and Wim Baert (Botanic Garden Meise) for their help with the molecular work, as well as Andreas Frisch and an anonymous referee for valuable comments on the manuscript.
Author ORCIDs
Damien Ertz, 0000-0001-8746-3187; André Aptroot, 0000-0001-7949-2594; Brian Coppins, 0000-0001-9464-0495.






