Introduction
Patterns of genetic variation among populations can reveal the evolutionary history of species. Population genetic differentiation and structure across a species’ distribution range is generally associated with the migration rates and its dispersal capability, but it could also be a product of demographic changes that occurred during the species colonization process into new habitats, such as population expansions or bottlenecks, which in turn mediate the effects of genetic drift and inbreeding (Carmichael et al., Reference Carmichael, Krizan, Nagy, Fuglei, Dumond, Johnson, Veitch, Berteaux and Strobeck2007; Latch et al., Reference Latch, Reding, Heffelfinger, Alcalá-Galván and Rhodes2014; Ngeve et al., Reference Ngeve, Van der Stocken, Menemenlis, Koedam and Triest2017). The combined action of genetic drift, selection and limited migration could result in local genetic variation, leading to genetic structure among populations (Hey and Machado, Reference Hey and Machado2003; Bradburd et al., Reference Bradburd, Ralph and Coop2013). In parasites, the population genetic composition is shaped not only by the parasite's intrinsic characteristics, such as transmission mode, number of hosts involved in the life cycle, and type of reproduction but also by certain host’ traits like population density, vagility, social structure and its particular population genetic makeup; each of these factors highly influence the microevolutionary processes of parasites (Nadler, Reference Nadler1995; Huyse et al., Reference Huyse, Poulin and Théron2005; Barrett et al., Reference Barrett, Thrall, Burdon and Linde2008; Blasco-Costa and Poulin, Reference Blasco-Costa and Poulin2013). Since many of the genetic attributes observed in parasite populations are driven by their host's demographic and dispersal histories, host and parasite phylogeographic patterns are expected to be intertwined, especially in highly host-specific parasites.
Pinworms are parasitic nematodes, directly transmitted and commonly found in primates (Hugot et al., Reference Hugot, Gardner and Morand1996). These nematodes are highly host specific, with one genus of pinworms parasitizing each major group of primates (Enterobius in Catarrhini; Lemuricola in Strepsirrhini; and Trypanoxyuris in Platyrrhini) and one to two species of pinworms specific to each host species, forming tight co-evolutionary associations with their primate hosts (Hugot, Reference Hugot1999). Trypanoxyuris is a genus of pinworms that infect Neotropical non-human primates; this genus currently includes 22 described species (Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez-Ponce de León2016).
Mantled howler monkeys (Alouatta palliata) and Central American spider monkeys (Ateles geoffroyi) inhabit tropical forests across Middle America, from the western coast of northern Peru, Ecuador and north-western Colombia to south-eastern Mexico (Rylands et al., Reference Rylands, Groves, Mittermeier, Cortes-Ortiz, Hines, Estrada, Garber, Pavelka and Luecke2006). Morphological and genetic variations of these primates are found along their range, with 5 recognized subspecies of Al. palliata (Alouatta palliata mexicana; Alouatta palliata palliata; Alouatta palliata aequatorialis; Alouatta palliata coibensis; Alouatta palliata trabeata) and allegedly 7 subspecies of At. geoffroyi (Ateles geoffroyi vellerosus; Ateles geoffroyi yucatanensis; Ateles geoffroyi geoffroyi; Ateles geoffroyi frontatus; Ateles geoffroyi ornatus; Ateles geoffroyi grisescens; Ateles geoffroyi azuerensis) (Rylands et al., Reference Rylands, Groves, Mittermeier, Cortes-Ortiz, Hines, Estrada, Garber, Pavelka and Luecke2006); all of them inhabiting different biogeographic areas across Mexico and Central America (Ford, Reference Ford, Estrada, Garber, Pavelka and Luecke2006) (distribution shown in Supplementary Material S1). A recent assessment of the phylogenetic relationships among Mesoamerican spider monkeys showed that the recognized subspecies of At. geoffroyi were not monophyletic, suggesting the need for a taxonomic revision of this group (Morales-Jimenez et al., Reference Morales-Jimenez, Cortés-Ortiz and Di Fiore2015).
Both species of primates are parasitized by two species of Trypanoxyuris; Trypanoxyuris minutus and Trypanoxyuris multilabiatus infect Al. palliata; while Trypanoxyuris atelis and Trypanoxyuris atelophora are found in At. geoffroyi (Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez Ponce de León2015, Reference Solórzano-García, Nadler and Pérez-Ponce de León2016). A study of the population genetics of T. minutus and T. atelis from primate populations inhabiting forest fragments across south-eastern Mexico showed genetic panmixia in both pinworm species despite habitat loss and fragmentation. This indicates that host isolation in time and space due to relatively recent changes in landscape configuration has not promoted genetic differentiation and structure among local parasite populations. The large population sizes of parasites could additionally be delaying the effects of genetic drift (Solórzano-García et al., Reference Solórzano-García, Gasca-Pineda, Poulin and Pérez-Ponce de León2017).
In this study, we expand the scale of the analysis by exploring a broader geographic range; pinworm specimens were sampled from Central American primate populations in Nicaragua and Costa Rica. Based on the genetic patterns observed in the parasite populations in light of the host phylogeography, we examined whether parasites from Central America represent the same evolutionary significant unit as the ones previously sampled in Mexico and whether biogeographic history and speciation of the primates explain the genetic variation of these parasites. To test our hypotheses, we compared the genetic divergence and phylogenetic history of four Trypanoxyuris species infecting different subspecies of howler and spider monkeys inhabiting tropical forests in Mexico, Nicaragua and Costa Rica. We followed the traditional classification for At. geoffroyi considering At. g. vellerosus and At. g. yucatanensis as separate subspecies, in order to test if parasite molecular data could serve as additional information to clarify the taxonomic status of these subspecies.
Materials and methods
Collection of specimens
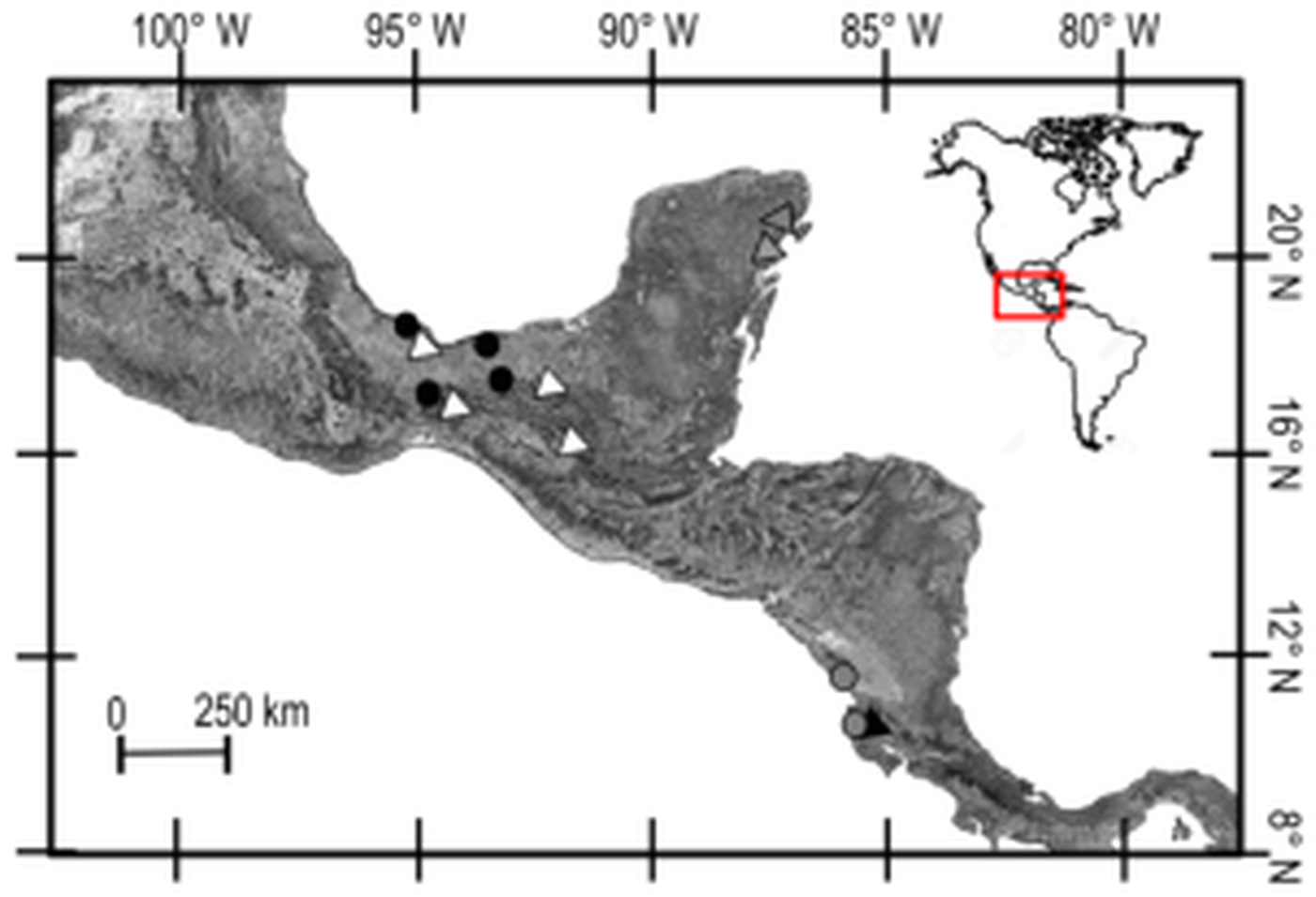
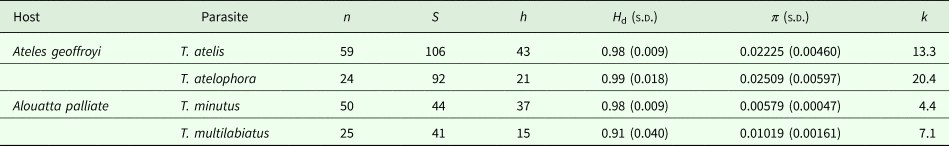
Trypanoxyuris specimens were collected from free-living populations of two subspecies of howler monkeys and three subspecies of spider monkeys. Alouatta palliata mexicana was sampled in four different locations in south-eastern Mexico, and Al. p. palliata was sampled in Apoyo, Nicaragua and Sector Santa Rosa, Guanacaste, Costa Rica (Fig. 1). Samples from At. geoffroyi vellerosus and At. g. yucatanensis were obtained in four and two south-eastern Mexico locations, respectively, and samples from At. g. frontatus were collected at Sector Santa Rosa, Guanacaste, Costa Rica (Fig. 1). Adult pinworms were recovered from faeces of these primates in situ and fixed either in 100% alcohol for DNA extraction, or 4% formalin for morphological examinations. Faecal samples were also collected and preserved at −4 °C to be examined for additional adult pinworms in the laboratory following the procedure suggested by Hasegawa (Reference Hasegawa, Huffman and Chapman2009). For morphological examination, worms were cleared with an alcohol-glycerol solution, and observed using an Olympus BX51 light microscope equipped with differential interference contrast. En face observations were made following the technique proposed by Hasegawa et al. (Reference Hasegawa, Ikeda, Dias-Aquino and Fukui2004). Finally, some specimens were processed for scanning electron microscopy (SEM) (Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez Ponce de León2015, Reference Solórzano-García, Nadler and Pérez-Ponce de León2016).
Fig. 1. Collection sites of Trypanoxyuris spp. from different host subspecies. Triangles = Ateles geoffroyi; black: At. g. frontatus; white: At. g. vellerosus; gray: At. g. yucatanensis. Circles = Alouatta palliata; black: Al. p. mexicana; gray: Al. p. palliata.
DNA extraction and sequencing
Individual pinworms fixed in ethanol were digested overnight at 56 °C in a solution containing 10 mm Tris-HCl (pH 7.6), 20 mm NaCl, 100 mm EDTA (pH 8.0), 1% Sarkosyl and 0.1 mg mL−1 proteinase K. DNA was extracted from the supernatant using the DNAzol® reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's instructions. A fragment of the mitochondrial cytochrome c oxidase subunit 1 gene (cox1), and a region of the large subunit of the nuclear ribosomal gene (28S) were amplified by polymerase chain reaction (PCR), using the procedure and primers specified in Solórzano-García et al. (Reference Solórzano-García, Nadler and Pérez-Ponce de León2016, Reference Solórzano-García, Gasca-Pineda, Poulin and Pérez-Ponce de León2017). Contig sequences of cox1 were aligned using CLUSTAL W and MESQUITE v. 2.75; two final sets of cox1 alignments were employed, one including all Trypanoxyuris species with available molecular information, consisting of 61 sequences with 707 bp, with 4–12 sequences per Trypanoxyuris species per host subspecies; and a species-specific alignment for each Trypanoxyuris species including all the previously published sequences of pinworms from primates in Mexico (Solórzano-García et al., Reference Solórzano-García, Gasca-Pineda, Poulin and Pérez-Ponce de León2017). The 28S contig sequences were aligned using MUSCLE and consisted of 35 sequences of 1149 bp, with 2–5 sequences per Trypanoxyuris species per host subspecies. All sequences obtained in this study were deposited in GenBank (28S: MH733397-MH733410; cox 1: MH733411–MH733440).
Phylogenetic analyses
Phylogenetic analyses were conducted through Maximum likelihood (ML) and Bayesian Inference (BI) separately for each gene. To add phylogenetic context, we included DNA sequences available in GenBank from other pinworm species from primates. Phylogenetic analyses for the cox1 alignments where conducted independently for each Trypanoxyuris species to corroborate the distinction between pinworm populations from different host subspecies. MrModeltest v.2.3 (Nylander, Reference Nylander2004) was used to select the best model of evolution for each gene using the AIC, selecting the GTR + I + G substitution model as the best model for both genes. ML trees were inferred using the program RaxML v.8 (Stamatakis, Reference Stamatakis2014) as implemented in the CIPRES Science Gateway (Miller et al., Reference Miller, Pfeiffer and Schwartz2010), using 1000 replications of bootstrap resampling to assess clade support. BI analyses were performed using MrBayes v.3.2.2 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003) and the CIPRES Science Gateway. Bayesian analyses included two simultaneous runs of MCMC, each for four million generations, sampling trees every 4000 generations, a heating parameter value of 0.2, and a ‘burn-in’ of 25%. A 50% majority-rule consensus tree was constructed from the post-burn-in trees.
Analyses of genetic variation
Median-joining cox1 haplotype networks were constructed using Network v.5.000 (Bandelt et al., Reference Bandelt, Forster and Röhl1999) independently for each Trypanoxyuris species to show the evolutionary relationships among haplotypes from pinworms infecting different primate subspecies. In the cases where the resulting networks were too complex, involving multiple alternative linkages, we used the Maximum Parsimony calculation post-processing option to visualize the most parsimonious tree (Polzin and Dabeschmand, Reference Polzin and Dabeschmand2003).
Genetic divergence in cox1 (P-distance) was calculated using MEGA v.6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013); standard error of the distances was estimated by bootstrap resampling with 500 replications. Molecular diversity indices including the number of segregating sites (S), number of haplotypes (h), haplotype diversity (H d), nucleotide diversity (π), and average number of nucleotide differences (k) were derived for cox1 using DnaSP v.5 (Rozas et al., Reference Rozas, Sánchez-DelBarrio, Messeguer and Rozas2003) for each species of Trypanoxyuris.
Results
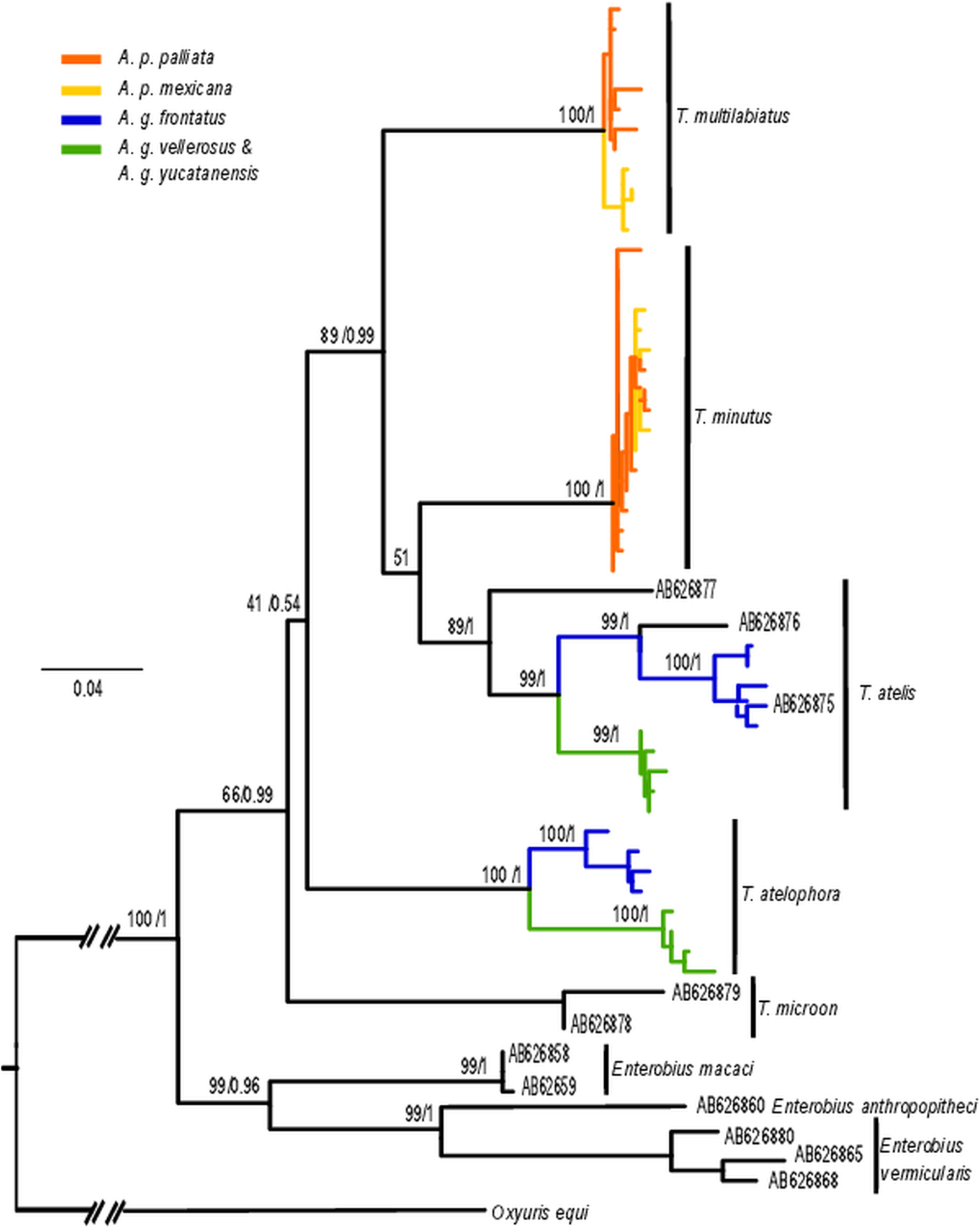
A pattern of genetic differentiation in concordance with host subspecies was evident in three of the four Trypanoxyuris species analysed (Fig. 2). Both the combined and the species-specific cox1 phylogenetic analyses showed that T. multilabiatus from the two Al. palliata subspecies (Al. p. mexicana and Al. p. palliata) formed separated clades; however, in T. minutus no pattern of genetic variation related to host subspecies was observed (Fig. 2). Similarly, cox1 sequences showed a pattern of differentiation between pinworms infecting different Ateles geoffroyi subspecies. Trypanoxyuris atelis and T. atelophora from At. g. frontatus sampled in Costa Rica grouped in clades separated from those pinworms collected from At. g. vellerosus and At. g. yucatanensis from Mexico (Fig. 2).

Fig. 2. Maximum likelihood cox1 phylogenetic tree of Trypanoxyuris spp. Numbers at the nodes represent ML bootstrap percentage, followed by posterior probabilities from Bayesian Inference. Trypanoxyuris species are indicated by bars at the right extreme of the tree. Trypanoxyuris from different host subspecies are marked as indicated at the top left of the figure. GenBank accession numbers indicate pinworm sequences acquired from different hosts: AB626877 Lagothrix lagotricha; AB626976 Ateles belzebuth; AB626875 Ateles geoffroyi; AB626878–79 Aotus azarae; AB626858–59 Macaca fuscata; AB626860, AB626880 Pan troglodytes; AB626865, AB626868 Homo sapiens.
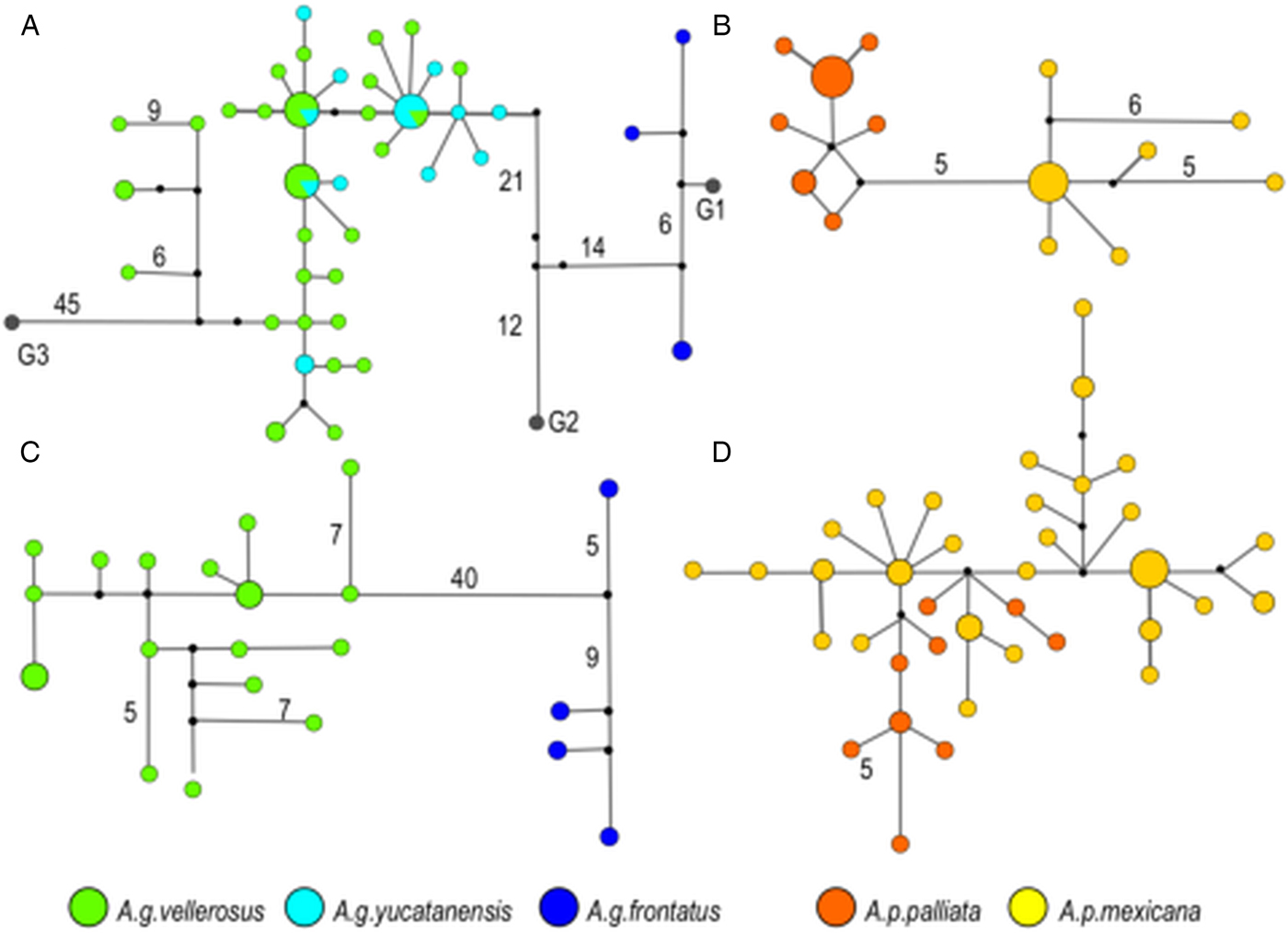
This pattern of genetic differentiation between host subspecies was also supported by the haplotype networks. In T. multilabiatus, T. atelis and T. atelophora median-joining networks showed separation of several mutations between pinworm haplotypes found in host subspecies from Mexico and Central America (Fig. 3). Haplotype separation between host subspecies was not observed in T. minutus (Fig. 3D), nor in T. atelis from At. g. vellerosus and At. g. yucatanensis (Fig. 3A). No shared haplotypes were found between pinworm populations infecting different host subspecies except in T. atelis from At. g. vellerosus and At. g. yucatanensis. Also, an ancestral haplotype from which the rest of the haplotypes are derived was not evident in any network (Fig. 3).
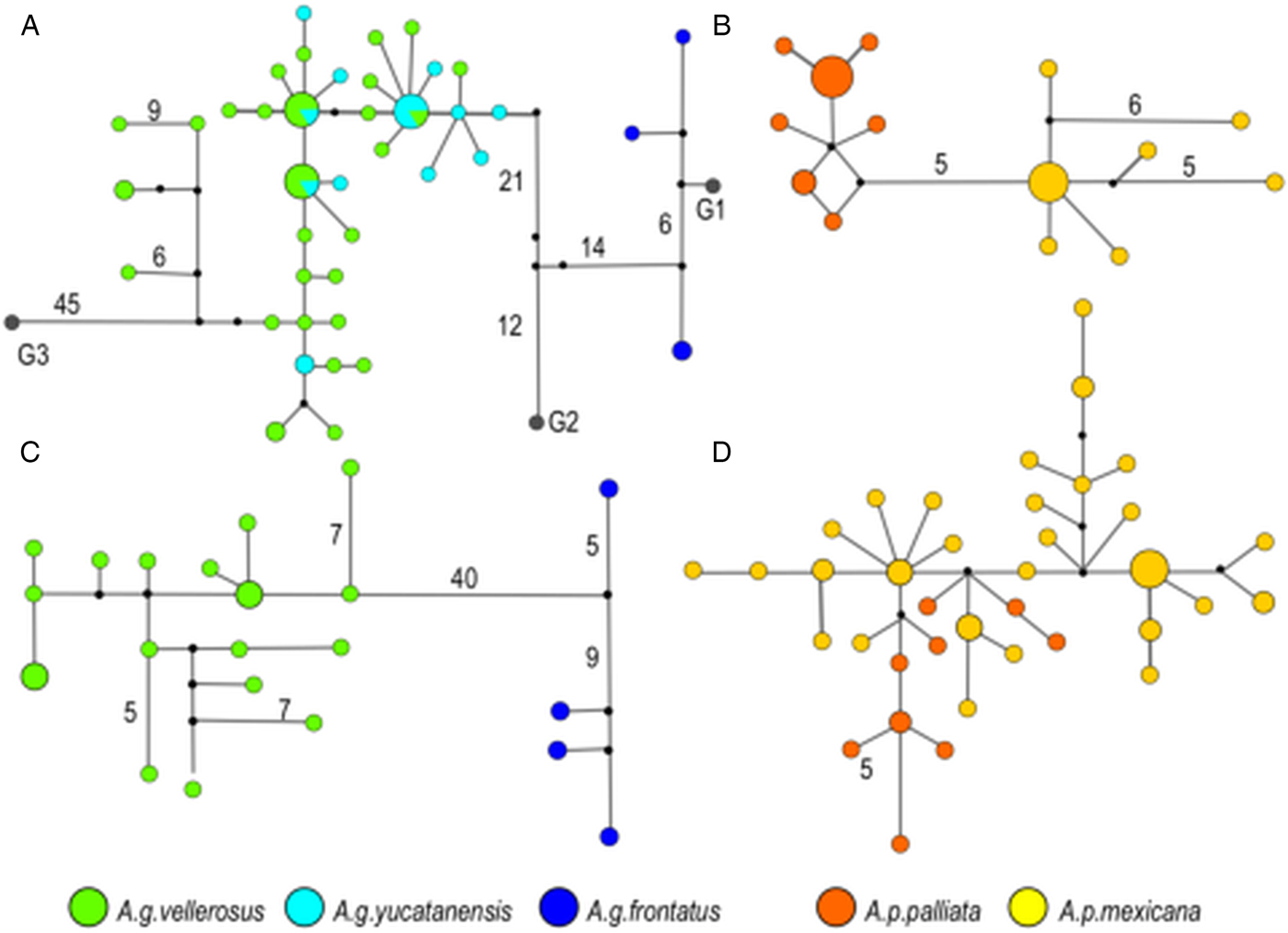
Fig. 3. Median-joining haplotype network based on cox 1 for Trypanoxyuris spp. Haplotype frequency is represented by the diameter of the circle. Numbers indicate mutational steps larger than 3 between haplotypes. (A) Trypanoxyuris atelis; (B) T. multilabiatus; (C) T. atelophora; (D) T. minutus. Trypanoxyuris from different host subspecies are marked as indicated at the bottom of the figure.
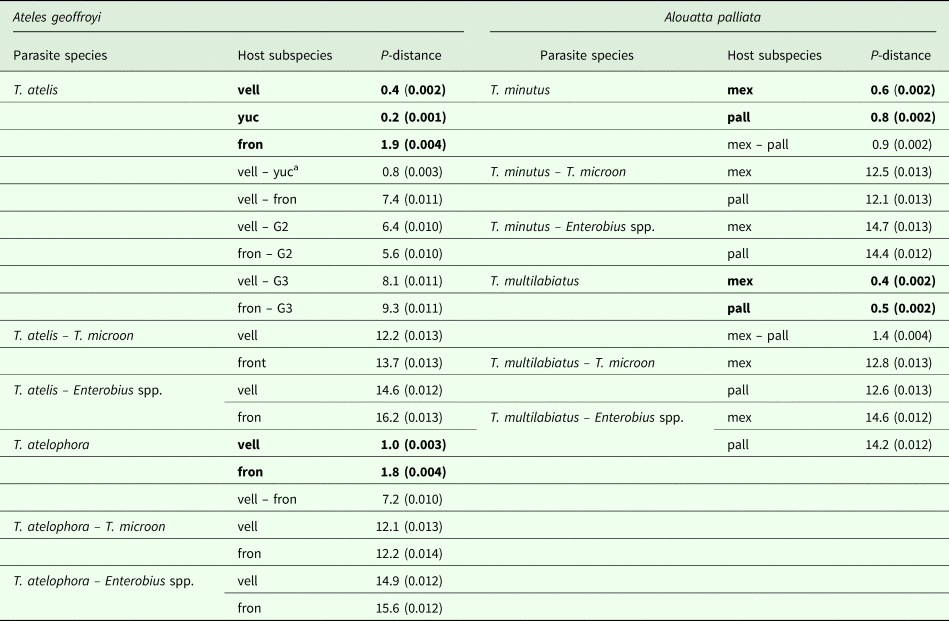
The genetic divergence estimated for cox1 among Trypanoxyuris parasitizing different primate subspecies ranged from 0.8% to 7.4%; the divergence among pinworm species ranged from 9.2% to 13.4% and from 14.2% to 16.2% between pinworm genera (Table 1). Within clades, the genetic divergence was very low and ranged from 0.8% in T. minutus to 1.9% in T. atelophora (Table 1). Genetic distances found among and within Trypanoxyuris clades from A. palliata are lower than the genetic distances found among and within Trypanoxyuris species sampled from A. geoffroyi (Table 1). All four Trypanoxyuris species are highly genetically diverse for mitochondrial DNA (mtDNA); however, pinworm populations infecting At. geoffroyi showed higher genetic diversity than pinworm populations from A. palliata (Table 2). In contrast, 28S rDNA sequences were identical within Trypanoxyuris species regardless of host subspecies.
Table 1. Cox1 genetic divergence among pinworms from different host subspecies
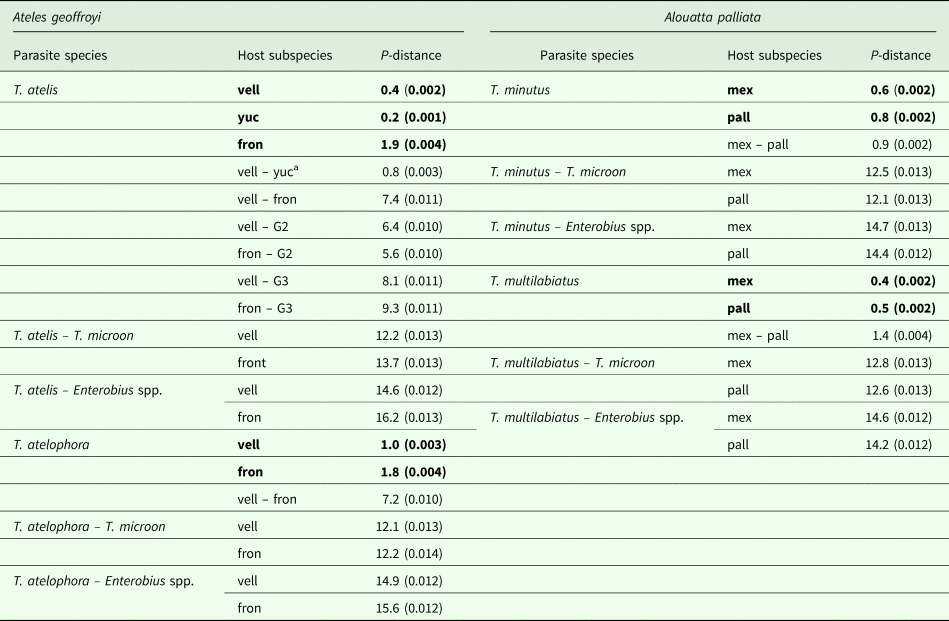
vell, vellerosus; yuc, yucatanensis; fron, fontatus; mex, mexicana; pall, palliata; G2, T. atelis from At. belzebuth (GenBank AB626876); G3, T. atelis from Lagothrix lagotricha (GenBank AB626877).
P distance expressed as percentage (± s.e.). Bold values indicate within clade distances.
a The genetic divergence between pinworms from yuc and the rest of the populations is not shown given the level of relatedness with those from vell, in order to avoid redundancy.
Table 2. Estimates of genetic diversity of the mitochondrial cox1 gene for each species of Trypanoxyuris collected from each primate host
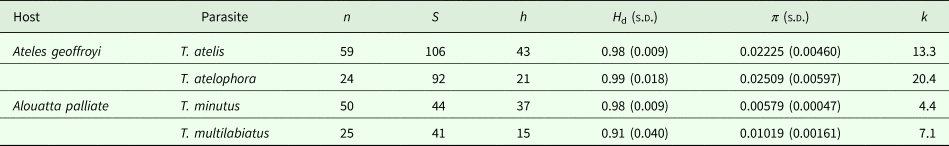
n, number of sequences; S, segregating sites; h, number of haplotypes; H d, haplotype diversity; π, nucleotide diversity; k, average number of nucleotide differences.
Despite the high levels of mitochondrial genetic variation observed between pinworms infecting different At. geoffroyi subspecies, no morphological differences were identified among them, either in males, females or in eggs, with diagnostic traits in agreement with the descriptions of T. atelis and T. atelophora (Hasegawa et al., Reference Hasegawa, Ikeda, Dias-Aquino and Fukui2004; Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez Ponce de León2015).
Discussion
Pinworms are highly host specific parasites that form strong co-evolutionary associations with their primate hosts (Hugot, Reference Hugot1999). In this study, we evaluated the population genetics of four Trypanoxyuris species infecting different howler and spider monkey subspecies distributed across Middle America and found genetic variation concordant with parasite–host association patterns.
As expected, morphological and molecular evidence indicate that the same species of Trypanoxyuris previously reported in Al. palliata and At. geoffroyi in Mexico (Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez Ponce de León2015, Reference Solórzano-García, Nadler and Pérez-Ponce de León2016) are also found parasitizing these primate species across its distribution in Central America, supporting the notion of the presence of two pinworm species per host species (Conga et al., Reference Conga, Giese, Serra-Freire, Bowler and Mayor2016; Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez-Ponce de León2016). Sequences of the 28S gene from primate populations from Central America and southern Mexico were identical within pinworms species regardless host subspecies, and no differences were observed through the morphological examination of the specimens with light and SEM. Nonetheless, phylogenetic analyses, haplotype relationships and genetic divergence in the mitochondrial gene cox1 showed some level of segregation of pinworm populations according to host subspecies for three of the four Trypanoxyuris species studied. The number of mutational steps estimated between haplogroups and the absence of shared haplotypes between Trypanoxyuris from different host subspecies suggest that local selective pressures related to each host subspecies along with restricted gene flow and the action of local genetic drift processes could have contributed to the differentiation of pinworm populations.
It is possible to speculate that the biogeographic history of these primates may have influenced the evolutionary dynamics among pinworm populations. Strong evidence suggests multiple independent waves of colonization of Central America by primates from northern South America throughout a series of founder events (Ellsworth and Hoelzer, Reference Ellsworth, Hoelzer, Lehman and Fleagle2006; Ford, Reference Ford, Estrada, Garber, Pavelka and Luecke2006). These independent colonization events and demographic fluctuations caused the intraspecific patterns of genetic and phenotypic variation currently observed across Al. palliata and At. geoffroyi range that justifies the designation of different populations as distinct subspecies. The patterns of genetic variation found among pinworm populations from different host subspecies are well explained by these historical events resulting in the genetic structure among howler and spider monkeys populations as well as their pinworms.
Both the genetic diversity and the level of genetic divergence found in Trypanoxyuris infecting spider monkeys (At. geoffroyi) was higher than the values observed in Trypanoxyuris from howler monkeys. The same genetic patterns of the parasites have been also observed in their hosts, with howler monkeys, Al. palliata tending to be less genetically diverse across its geographical range than the spider monkeys, At. geoffroyi (Jasso-del Toro et al., Reference Jasso-del Toro, Márquez-Valdelamar and Mondragón-Ceballos2016; Ruiz-García et al., Reference Ruiz-García, Lichilín, Escobar-Armel, Rodríguez, Gutiérrez-Espeleta, Ruiz-García and Shostell2016, Reference Ruiz-García, Cerón, Sánchez-Castillo, Rueda-Zozaya, Pinedo-Castro, Gutierrez-Espeleta and Shostell2017). Moreover, the genetic distinctiveness of Al. palliata subspecies uncovered through mtDNA is relatively minor (0.5%) (Cortés-Ortiz et al., Reference Cortés-Ortiz, Bermingham, Rico, Rodriguez-Luna, Sampaio and Ruiz-Garcia2003), while that reported for At. geoffroyi subspecies is ~5% (Collins and Dubach, Reference Collins and Dubach2000).
The two pinworm species found in Al. palliata showed contrasting results in the population genetics and phylogenetic analyses; the genetic differentiation of T. multilabiatus was closely tied with howler monkey subspecies, while the species T. minutus formed one single clade with no structure in terms of host subspecies. This disparity on the genetic configuration could be explained by the population size of each pinworm species, and by the strength of the evolutionary ties between host and parasite given by the level of host specificity. Trypanoxyuris multilabiatus has only been reported in Al. palliata (Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez-Ponce de León2016), whereas T. minutus is a common and abundant pinworm reported in several species of howler monkeys along their distributional range in the Neotropical biogeographic region; i.e. Al. guariba, Al. seniculus and Al. cayara (Solórzano-García and Pérez-Ponce de León, Reference Solórzano-García and Pérez-Ponce de León2018). Hence, T. minutus can be regarded as a more generalist parasite species, and perhaps less susceptible than T. multilabiatus to subtle changes in the physiology, genetics and demography linked to host subspecies. Furthermore, T. multilabiatus showed the lowest haplotype diversity, probably as a consequence of a small population size that could be related to the series of population reductions experienced by A. palliata during its dispersal through Central America up to south-eastern Mexico (Cortés-Ortiz et al., Reference Cortés-Ortiz, Bermingham, Rico, Rodriguez-Luna, Sampaio and Ruiz-Garcia2003; Ford, Reference Ford, Estrada, Garber, Pavelka and Luecke2006), whereas a broader host distribution and hence, larger population sizes could have allowed the other species, T. minutus to cope with such demographic changes in the host.
Both pinworm species found in spider monkeys, T. atelis and T. atelophora, showed genetic differences associated with host subspecies, the two alleged subspecies occurring in Mexico (At. g. vellerosus and At. g. yucatanensis) and the subspecies from Central America (At. g. frontatus). However, no separation was observed between T. atelis from At. g. vellerosus and At. g. yucatanensis. Molecular phylogenetic analyses of Central American spider monkeys through a multilocus approach of mtDNA have shown a lack of distinction between At. g. vellerosus and At. g. yucatanensis, and thus they should be considered as the same subspecies, At. g. vellerosus (Morales-Jimenez et al., Reference Morales-Jimenez, Cortés-Ortiz and Di Fiore2015). The resulting genetic patterns of the parasites unveiled in this study constitute additional evidence supporting this proposal.
Our results showed substantial intraspecific genetic differences among cox1 sequences from T. atelis and T. atelophora collected from At. g. vellerosus and At. g. frontatus (7.4% and 7.2%, respectively). MtDNA is characterized as rapidly evolving among nematodes (Blouin, Reference Blouin1998); however, the genetic divergence observed between Trypanoxyuris populations from these two spider monkey subspecies is noticeably high considering the intraspecific genetic distances reported in pinworms. The mean intraspecific genetic divergence, for the same molecular marker, among species of Trypanoxyuris occurring in Mexico, varied between 0.5% and 1.1% (Solórzano-García et al., Reference Solórzano-García, Nadler and Pérez-Ponce de León2016), while the maximum intraspecific genetic distance in cox1 reported for pinworms is 6.5% in Enterobius vermicularis (Nakano et al., Reference Nakano, Okamoto, Ikeda and Hasegawa2006). The time at which At. g. vellerosus and At. g. frontatus diverged from the common ancestor has been estimated ~1.5 Ma (Morales-Jimenez et al., Reference Morales-Jimenez, Cortés-Ortiz and Di Fiore2015), and this might be considered a relatively recent date for the accumulation of the observed genetic differences in these pinworm populations. Moreover, specimens of T. atelis have been previously obtained and sequenced from different primate species of the family Atelidae, all from captive populations in Japan, including the spider monkey, At. geoffroyi (unknown subspecies), the yellow-bellied spider monkey, At. belzebuth, and the brown woolly monkey, Lagothrix lagotricha (Hasegawa et al., Reference Hasegawa, Sato and Torii2012) (Fig. 2). The ability of T. atelis to infect different host species in addition to the magnitude of the genetic variation among populations of this pinworm imply that the species of Trypanoxyuris in spider monkeys exhibit a high evolutionary potential. This would facilitate adaptation to the specific selective pressures of each host, resulting in the presence of different genetic configurations, but could also indicate the potential existence of cryptic species (Nadler and Pérez-Ponce de León, Reference Nadler and Pérez-Ponce de León2011) of Trypanoxyuris parasitizing spider monkeys.
The results of this study show how genetic patterns uncovered in parasitic organisms may reflect host phylogeny and biogeography (Whiteman and Parker, Reference Whiteman and Parker2005; Criscione, Reference Criscione2008). Moreover, because of the intimate association between pinworms and primates, genetic data of the parasite could shed light on host evolutionary history, helping us to better understand primate phylogenetic relationships and vice versa. These additional data aid the construction of a more accurate taxonomy. Still, denser taxon sampling and sequencing work are required to test the strength of the co-evolutionary processes between pinworms and primates.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182018001749.
Acknowledgements
We thank R. Blanco Segura and M. M. Chavarria and staff from the Área de Conservación Guanacaste and Ministerio de Ambiente y Energía, Costa Rica; Martha Barluenga for her support during fieldwork in Nicaragua; and all the Natural Protected Areas authorities, Arqueological Parks, and landowners that kindly granted permission for the collection of samples in Mexico. Monica Myers, Saul Cheves Hernández and Evin Murillo Chacon provided valuable assistance during fieldwork in Costa Rica. We also thank Laura M. Márquez for sequencing service.
Financial support
This study was partially funded by the Posgrado en Ciencias Biológicas UNAM through the programme Fomento a la Graduación Temprana to BSG, the Canada Research Chairs Program and a National Sciences and Engineering Council of Canada Discovery grant to ADM, and a Chester Zoo grant to FA.
Conflict of interest
None.
Ethical standards
Not applicable.