Introduction
Parasitic nematodes from fishes of the Indian subcontinent have been studied since the early 20th century (Baylis & Daubney, Reference Baylis and Daubney1922; Soota, Reference Soota1983; Sood, Reference Sood1988, Reference Sood2017), with over 600 species recorded to date. Of these, ascaridoids are reportedly represented by species in the following genera: Aliascaris Kalyankar, 1971; Alibagascaris Kalyankar, 1970; Hysterothylacium Ward & Magath, 1917; Iheringascaris Pereira, 1935; Lappetascaris Rasheed, 1965; Mehdiascaris Kalyankar, 1969; Paranisakis Baylis, 1923; Raphidascaris Railliet & Henry, 1915; and Raphidascaroides Yamaguti, 1914 (Soota, Reference Soota1983; Malta et al., Reference Malta, Paiva, Elisei, Tavares and Pereira2018, Reference Malta, Paiva, Elisei, Tavares and Pereira2020). During a long-term survey of parasites of fishes from West Bengal, worms belonging to a previously unknown species of ascaridoid were collected from the gastrointestinal tract of a unique fish host, Apocryptes bato (Hamilton, 1822) (Actinopterygii: Gobiidae: Oxydercinae), a mudskipper, and the only species in its monotypic genus (Murdy & Jaafar, Reference Murdy, Jaafar, Jaafar and Murdy2017). Apocryptes bato ranges from eastern India to Myanmar (Talwar & Jhingran, Reference Talwar and Jhingran1991; Barman et al., Reference Barman, Mukherjee and Kar2000; Parenti & Jaafar, Reference Parenti, Jaafar, Jaafar and Murdy2017), and is widely distributed in the central and southern parts of West Bengal. Despite its wide distribution, little is known about its parasite fauna. Except for a few myxozoans (Bajpai & Haldar, Reference Bajpai and Haldar1982) no parasite has previously been reported from this fish.
On closer examination, this ascaridoid was identified as an undescribed species of Raphidascaris. Species of Raphidascaris parasitize fishes in the fresh waters of North America, South America, Eurasia and Japan, as well as in marine environments (Soota, Reference Soota1983; Moravec, Reference Moravec1994; Moravec, Reference Moravec1998; Hoffman, Reference Hoffman1999; Moravec & Nagasawa, Reference Moravec and Nagasawa2002; Moravec & Justine, Reference Moravec and Justine2020). A few species of this genus have been reported from marine and freshwater fishes in South Asia, including India (Soota, Reference Soota1983; Sood, Reference Sood1988, Reference Sood2017). In this study, we describe and characterize this new species found in a freshwater fish, using morphological and molecular data, and discuss the taxonomic status of other Raphidascaris species in fishes from the Indian subcontinent.
Materials and methods
Collection and morphological study
From December 2007 to March 2020, 221 mudskippers (A. bato) were collected from the Mundeswari River in West Bengal, India. Live worms, isolated from digestive tract, were fixed in hot 4% formaldehyde solution and preserved in 70% ethanol (Moravec, Reference Moravec1994). A few worms were directly fixed in 100% undenatured ethanol for subsequent DNA extraction and amplification. Nematodes were cleared using glycerine or lactophenol for light microscopical examination, using an Olympus BX53 microscope (Olympus corporation, Tokyo, Japan). Drawings were made with an Olympus BX53 drawing attachment. For scanning electron microscopy (SEM), specimens were post fixed in 1% osmium tetroxide, dehydrated with a graded alcohol series, infiltrated with hexamethyldisilazane and air-dried (modified from Bowen et al., Reference Bowen, Hemann, Johnson and Good1990). The specimens were then coated with gold and examined with a Zeiss Sigma-300 FE Scanning Electron Microscope (Carl Zeiss AG, Oberkochen, (Baden-Württemberg), Germany) at an accelerating voltage of 10 KV or with a Hitachi TM 3030+ benchtop Scanning Electron Microscope (Hitachi Ltd., Tokyo, Japan) at an accelerating voltage of 15 KV. All measurements are in micrometres unless otherwise stated.
Scientific and common names of fishes follow Froese & Pauly (Reference Froese and Pauly2019). Specimens are being deposited in the following museum collections: Zoological Survey of India, Kolkata, India (ZSI); the Harold Manter Laboratory of Parasitology (HWML), University of Nebraska, Lincoln, Nebraska, USA; and in the Helminthological Collection of the Institute of Parasitology of the Biology Centre of the Czech Academy of Sciences in České Budějovice, Czech Republic (IPCAS).
Molecular work and phylogenetic analysis
A small (~5–8 mm) portion was excised from the mid region of two adult worms (male and female) that had been stored in 100% undenatured ethanol, and genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, California, USA). The remaining corresponding portions of the worms were stored in 95% or 100% ethanol as vouchered ‘hologenophores’ (sensu Pleijel et al., Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008). The 28S ribosomal RNA (rRNA) gene was amplified using the following primers; 391a (forward) 5′-AGCGGAGGAAAAGAAACTAA-3′, and 501 (reverse); 5′-TCGGAAGGAACCAGCTACTA-3′ (Carreno & Nadler, Reference Carreno and Nadler2003). Polymerase chain reaction (PCR) reactions were performed on an Applied Biosystems Veriti thermal cycler (Applied Biosystems, Thermo Fisher Scientific, Waltham, Massachusetts, USA) using 0.25–0.5 μl of Ex Taq DNA polymerase (TaKaRa Bio USA, Inc., Mountainview, California, USA) in a total reaction volume of 50 μl containing 31.75 μl of nuclease-free water (Qiagen Inc.), 5 μl of extracted DNA as template, 2 μl each of forward and reverse primers at a concentration of 1 pmol/μl, 5 μl of 10X Ex Taq Buffer (Mg2+ plus) and 4 μl (200 μM) of dNTPs (deoxynucleoside triphosphates) (TaKaRa Bio USA, Inc.).The amplification protocol consisted of an initial denaturing cycle of 5 min at 94°C, 25–35 cycles of the following: 94°C for 30 s, 54°C for 30 s, 72°C for 1 min and a final elongation at 72°C for 5 or 7 min. PCR products were purified using ExoSAP-IT Express PCR Product Cleanup (Affymetrix Inc., Santa Clara, California, USA). Purified products were sent to MCLab (South San Francisco, California, USA) for automated sequencing. The PCR amplification primers and the following internal primers were used for sequencing: 500 (forward) 5′-ACTTTGAAGAGAGAGTTCAAGAG-3′, 503 (reverse) 5′-CCTTGGTCCGTGTTTCAAGACG-3′ and 504 (forward) 5′-CAAGTACCGTGAGGGAAAGTTG-3′ (Nadler et al., Reference Nadler, D'Amelio, Fagerholm, Berland and Paggi2000; Carreno & Nadler, Reference Carreno and Nadler2003; Nadler Lab UC Davis databases: https://nadlerlab.faculty.ucdavis.edu/lab-protocols-and-databases/).
Contigs were manually checked, edited for accuracy and trimmed using FinchTV (Geospiza Inc., Seattle, Washington, USA), and assembled in MEGA 7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). A 1005 bp consensus sequence from one worm was generated after assembling the contigs and used for the analysis. This sequence was aligned with sequences of 25 ascaridoids belonging to Raphidascarididae and one sequence each of Contracaecum multipapillatum (Drasche, 1882) (AF226574), Anisakis sp. (AY821759) and Heterocheilus tunicatus Diesing, 1839 (AF226592) available in GenBank, using Clustal W in MEGA 7. The last three taxa were included because two of them, C. multipapillatum and Anisakis sp., represent Anisakidae, a family related to Raphidascarididae, and H. tunicatus is an earlier branching lineage suitable for rooting the tree (Nadler & Hudspeth, Reference Nadler and Hudspeth2000; Nadler et al., Reference Nadler, D'Amelio, Fagerholm, Berland and Paggi2000). The final 28S rDNA aligned and trimmed dataset of these 29 sequences contained 444 positions. The markedly unequal coverage of the 28S rRNA gene by these sequences resulted in this, much smaller, number of positions in the final dataset. The sequence of the ascaridoid generated in this study was deposited in GenBank (www.ncbi.nlm.nih.gov) with the following accession number: MZ611858.
The aligned 28S rDNA sequence dataset was analysed using the Maximum Likelihood (ML) method and the Hasegawa–Kishino–Yano (HKY) model of substitution in MEGA 7. HKY + G+I – that is, the HKY model with a discrete gamma distribution (G) and allowing for some sites to be evolutionarily invariable (I) – was determined to be the appropriate model of substitution for the analysis, by testing for best fit using the algorithm implemented under ‘Test Model’ in MEGA 7. The 28S rDNA dataset was also analysed using Bayesian inference (BI) (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001) executed with Mr Bayes in Geneious Prime, version 2021.1.1, and through the Cyberinfrastructure for Phylogenetic Research (CIPRES) supercomputer Portal (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). Bayesian posterior probability values were determined after running the Markov chains (two runs, four chains) for four million generations and discarding the initial one-fourth of sampled trees as burn-in, with trees sampled every 4000 generations. The analysis involved 29 nucleotide sequences.
Results
Raphidascaris mundeswariensis n. sp. (Nematoda: Ascarida: Raphidascarididae)
Description
General. Medium-sized worms with thin cuticle; flaccid when collected live, maximum width at the posterior region of oesophagus. Anterior end with three well-developed lips, dorsal lip shorter than ventrolateral lips (figs 1a–c and 2a, b). Lips without lateral membranous flanges but their oral edges slightly set off by narrow borders with irregular margins. Dorsal lip with two subdorsal double papillae (figs 1c and 2b), each ventrolateral lip with one double papilla, one single papilla and one amphid situated laterally (figs 1a, c and 2a–c). Interlabia and lateral alae absent. Intestinal caecum absent. Oesophagus short; broader posteriorly than anteriorly. Excretory pore posterior to nerve ring (fig. 1a, b). Ventriculus relatively short and broad. Ventricular appendix prominent and moderately robust, ventral in position (fig. 1a, b). Tails of both sexes relatively short, conical in shape, without discernible spines (figs 1d, f, g and 2e, g).

Fig. 1. Raphidascaris mundeswariensis n. sp.: (a) anterior region of female; (b) anterior region of male; (c) female, apical (en face) view; (d) caudal region of male, lateral view; (e) vulval region of female, dorsolateral view; (f) posterior end of female, lateral view; (g) posterior end of male, subventral view.
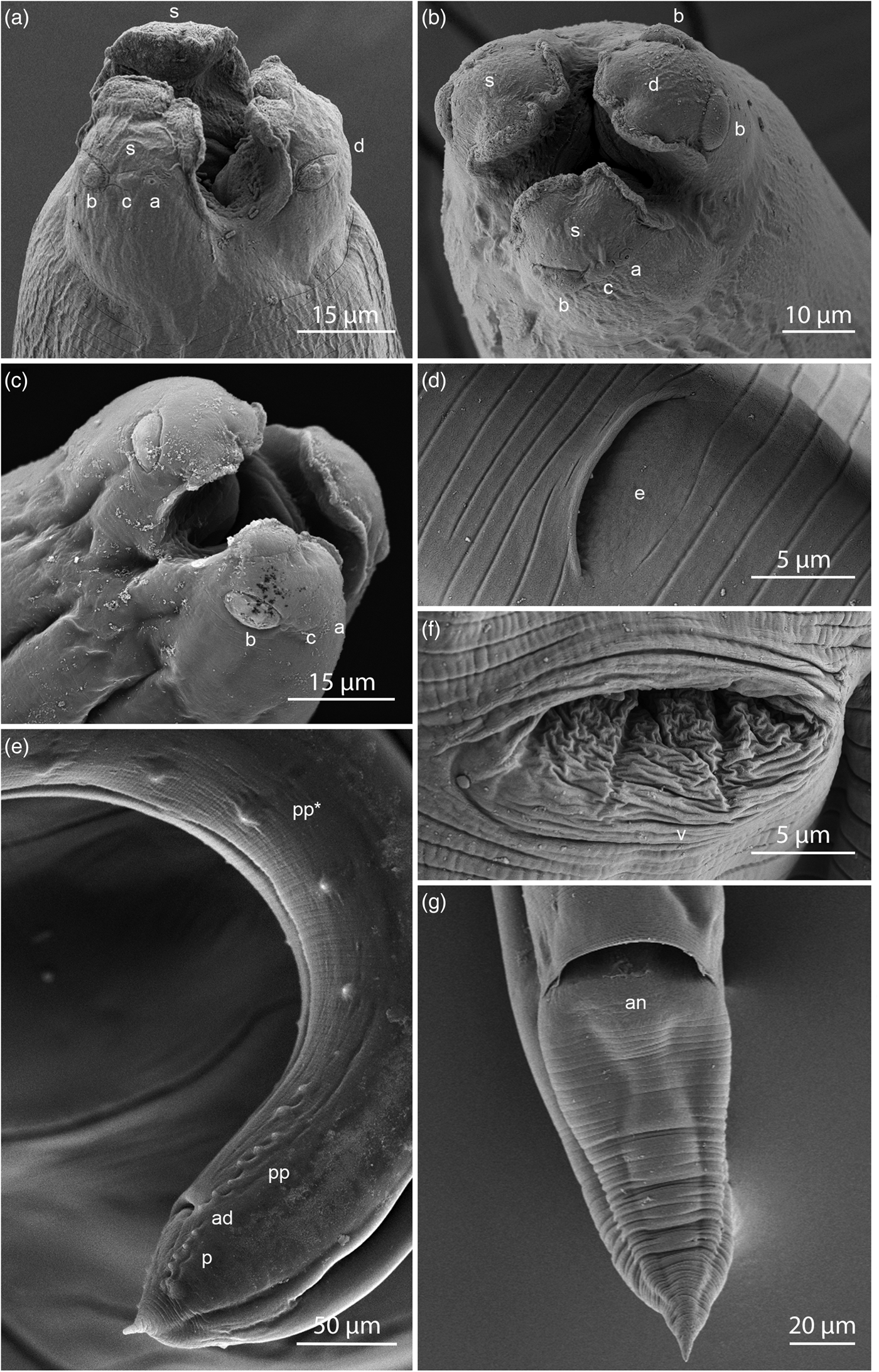
Fig. 2. SEM micrographs of Raphidascaris mundeswariensis n. sp. showing (a) female, cephalic end, oblique apical view of subventral and dorsal lips view; (b) female, apical view; (c) male, cephalic end, oblique apical view; (d) excretory pore of female; (e) caudal region of male, left ventrolateral view; (f) vulva of female; (g) tail of female, ventral view. Abbreviations: a, amphid; ad, adanal papillae; an, anus; b, double papilla; c, single papilla; d, dorsal lip; e, excretory pore; p, postanal papillae; pp, smaller preanal papillae; pp*, larger preanal papillae; s, subventral lip; v, vulva.
Male. Based on six mature specimens; measurements of the holotype in parentheses. Body length 3.73–5.44 (4.04) mm, maximum width at the end of oesophagus 106–202 (112). Dorsal lip 35–48 (39) long, 30–42 (39) wide. Right ventrolateral lip 39–52 (42) long, 21–30 (22) wide. Left ventrolateral lip 37–55 (45) long, 23–33 (25) wide. Oesophagus 419–562 (433) long (8.9–11.1% of total body length), 73–125 (73) wide near club-shaped base. Nerve ring and excretory pore 144–221 (152) and 306–381 (319), respectively, from anterior end. Nerve ring encircling oesophagus at end of first 26.87–30.36% of oesophageal length. Ventriculus 42–79 (42) long, 81–99 (83) wide. Ventricular appendix 219–339 (223) long, 29–63 (45) wide. Spicules equal, tapering to a pointed end, 214–255 (216) long, representing 17.05–22.17% of body length (fig. 1d). Gubernaculum absent. Distance of single testes loop from anterior end 571–913 (738). Seminal vesicle 706–1287 (772) long, 79–148 (91) wide. Ejaculatory duct 494–844 (506) long, 31–93 (31) wide. Caudal papillae arranged as follows; 14 pairs of preanal papillae of two markedly different sizes, one set of 6–7 smaller preanal papillae closer to the anus and 7–8 larger, more widely spaced preanal papillae, one pair of adanal papillae and six pairs of postanal papillae (figs 1d and 2e). Tail (from anus to tip) 69–104 (72) long, 69–106 (69) wide at anus (figs 1d, g and 2e). Caudal mucron 15–19 (16) long.
Female. Based on seven mature specimens; measurements of the allotype in parentheses. Body length 7.06–11.92 (7.06) mm, width at end of oesophagus 155–454 (155). Dorsal lip 39–76 (45) long, 48–62 (58) wide. Right ventrolateral lip 48–80 (48) long, 30–53 (30) wide. Left ventrolateral lip 52–73 (52) long, 34–48 (34) wide. Oesophagus 513–947 (513) long (9.01–13.04% of total body length), 94–228 (94) wide near club-shaped base. Nerve ring and excretory pore 175–288 (181) and 248–424 (248), respectively, from anterior end. Nerve ring encircling oesophagus at end of first 26.35–36.92% of oesophageal length. Ventriculus 44–94 (56) long, 78–157 (81) wide. Ventricular appendix 288–415 (325) long, 44–106 (50) wide. Vulva pre-equatorial, 956–1777 (956) from anterior end. Vagina 300–438 (381) long. Egg rounded to sub-oval, 19–29 (25) long, 36–44 (41) wide, terminal eggs containing two-celled embryo (fig. 1e). Anterior ovarian loop 556–994 (994) posterior to oesophagus; posterior ovarian loop 495–619 (572) anterior to anus, respectively. Tail (from anus to tip) 181–352 (181) long, 106–138 (106) wide at anus (figs 1f and 2g).
Taxonomic summary
Type host. Apocryptes bato (Hamilton, 1822) (Gobiidae: Oxydurcinae).
Site of infection. Intestine.
Type locality. Mundeswari River, Ranjitbati, Hooghly, West Bengal, India (22° 40' 59.6"N, 87° 53' 24.5"E).
Prevalence and intensity of infection. The prevalence of infection was 16.74% (from 221 mudskippers examined between December 2017 and March 2020). The mean intensity of infection was 2.32 (1–14 worms/host).
Specimens deposited. Holotype (male): ZSI/WN 3112; allotype (female): ZSI/WN 3113; hologenophore: HWML 112250; paratypes and vouchers: HWML 112247–112249; IPCAS N-1256; ZSI/WN 3114 and 3114/1.
Genetic data. 28S rRNA gene sequence (partial) (GenBank accession numbers MZ611858).
Etymology. The species name is derived from the Mundeswari River, the type locality of this parasite.
Molecular characterization and phylogenetic analyses
The ML phylogenetic tree (fig. 3) shows Raphidascaris mundeswariensis n. sp. in a strongly supported clade containing Raphidascaris gigi Fujita, Reference Fujita1928, Raphidascaris lophii Wu, Reference Wu1949, Raphidascaris longispicula Li, Liu & Zhang, 2012 and two species of Hysterothylacium. The analysis also suggests that neither Raphidascaris nor Hysterothylacium are monophyletic, although 11 of the 14 species of Hysterothylacium in the analyses formed three strongly supported separate clades with their congeners. The analysis also failed to recover Raphidascaroides as a monophyletic lineage, but Raphidascaroides moraveci and Raphidascaroides brasiliensis formed a strongly supported clade.

Fig. 3. Phylogenetic tree generated from the maximum likelihood analysis of partial 28S rRNA gene sequences of Raphidascaris mundeswariensis n. sp. and other species in Raphidascarididae. GenBank accession numbers follow taxa. The tree is drawn to scale with branch lengths measured in the number of substitutions per site (scale bar shows the number of substitutions per site), and bootstrap values (1000 replicates) are indicated at the nodes. For comparison, Bayesian Posterior Probability values from the BI tree (supplementary material S1) are indicated below the nodes of the clade containing the new species.
The BI tree (supplementary material S1) showed the same topology as the ML tree with the new species, R. mundeswariensis, in the same strongly supported clade as before. The interrelationships of the other raphidascaridids were also consistent with those found in the ML analysis.
Remarks
Phylogenetic analyses suggest that the new taxon is closely related to R. gigi, R. lophii and R. longispicula. Morphological comparisons of R. mundeswariensis n. sp. to these species reveal a general resemblance, but some distinct differences exist between these three species and the new taxon. Raphidascaris gigi possess 26–30 pairs of preanal, 2–3 pairs of adanal and nine pairs of postanal papillae in contrast to 14 pairs of preanal, one pair of adanal and six pairs of postanal papillae, respectively, in R. mundeswariensis. Raphidascaris gigi also possesses longer spicules (354–476 μm long, 5.4–6.7% of body length) than R. mundeswariensis (214–255 μm long, 17.05–22.17% of body length) (calculated from Moravec & Nagasawa, Reference Moravec and Nagasawa2002; this study). The new species can also be readily distinguished from R. lophii (26–32 pairs of preanal, 3–4 pairs of para-anal and 8–11 pairs of postanal papillae) and from R. longispicula (25–28 pairs of preanal, 1–2 pairs of para-anal and 6–8 pairs of postanal papillae). Raphidascaris lophii and R. longispicula also have longer spicules (490–882 μm and 1130–1320 μm, respectively) than R. mundeswariensis (214–255 μm). The absence of lateral alae in R. mundeswariensis n. sp. further differentiates it from R. lophii and R. longispicula, which possess alae (from Li et al., Reference Li, Liu, Liu and Zhang2012; Xu et al., Reference Xu, Zhang, Liu and Li2012). Furthermore, R. lophii and R. longispicula were described from marine fish hosts.
Excluding species of the subgenera Sprentascaris and Ichthyascaris, four valid species of Raphidascaris have been reported from freshwater hosts to date – namely, the type species, Raphidascaris acus (Bloch, 1779), and three others: Raphidascaris cyprini Wang, 1965, Raphidascaris leiocassis Wang, 1965 and R. gigi. Raphidascaris mundeswariensis n. sp. can be clearly distinguished from these species as follows: R. acus possesses 16–21 pairs of preanal, 1–2 pairs of adanal and four pairs of postanal papillae and lateral flanges on the lips (Smith, Reference Smith1984), R. cyprini Wang, 1965 possesses longer spicules (400 μm) and two pairs of postanal papillae and R. leiocassis Wang, 1965 possesses 23 pairs of preanal and seven pairs of postanal papillae (Wang, Reference Wang1965; Li et al., Reference Li, Gibson and Zhang2016). The differences with the fourth freshwater species, R. gigi, have been previously mentioned.
The presence of two discernible groups of preanal papillae, as observed in R. mundeswariensis n. sp., appears to be uncommon among raphidascaridids. This feature is present in the type species of Raphidascaris, R. acus (Smith, Reference Smith1984, Fig. 7), as well as in R. brasiliensis Moravec & Thatcher, Reference Moravec and Thatcher1997 and R. moraveci Pereira, Tavares, Scholz, & Luque, Reference Pereira, Tavares, Scholz and Luque2015 (Moravec, Reference Moravec1998; Pereira et al., Reference Pereira, Tavares, Scholz and Luque2015), but not in the three Raphidascaris species, R. gigi, R. longispicula and R. lophii, belonging to the same clade as R. mundeswariensis (Moravec & Nagasawa, Reference Moravec and Nagasawa2002; Li et al., Reference Li, Liu, Liu and Zhang2012; Xu et al., Reference Xu, Zhang, Liu and Li2012).
Discussion
The genus Raphidascaris Railliet & Henry, 1915, currently includes approximately 32 species that parasitize fishes of fresh, brackish and marine waters (Soota, Reference Soota1983; Smith, Reference Smith1984; Moravec, Reference Moravec1994; Moravec, Reference Moravec1998; Hoffman, Reference Hoffman1999; Moravec & Nagasawa, Reference Moravec and Nagasawa2002; Moravec & Justine, Reference Moravec and Justine2020) and is typified by the following characters: (1) ventricular appendix present but intestinal caecum absent; (2) gubernaculum absent; and (3) lips without postlabial ring or conical processes (Soota, Reference Soota1983; Li et al., Reference Li, An and Zhang2007).
The interrelationships of various ascaridoid taxa including the genus Raphidascaris are controversial, and the taxonomy and systematics of raphidascaridids are still in flux; the monophyly of Raphidascaris and Hysterothylacium remains doubtful (Malta et al., Reference Malta, Paiva, Elisei, Tavares and Pereira2018, Reference Malta, Paiva, Elisei, Tavares and Pereira2020; this study). The genus Raphidascaris was previously subdivided on morphological grounds into three subgenera – namely, Ichthyascaris Wu, Reference Wu1949, Raphidascaris Railliet & Henry, 1915 and Sprentascaris Petter & Cassone, 1984 (Moravec et al., Reference Moravec, Kohn and Fernandes1990; Moravec & Nagasawa, Reference Moravec and Nagasawa2002; Li et al., Reference Li, Liu, Liu and Zhang2012, Reference Li, Gibson and Zhang2016). Recent integrative taxonomic analyses have shown that Sprentascaris can be considered a separate valid genus (Malta et al., Reference Malta, Paiva, Elisei, Tavares and Pereira2018, Reference Malta, Paiva, Elisei, Tavares and Pereira2020), a conclusion supported by our analyses (see fig. 3).
Previous phylogenetic analyses of raphidascaridids varied in their taxon sampling (Pereira et al., Reference Pereira, Tavares, Scholz and Luque2015; Malta et al., Reference Malta, Paiva, Elisei, Tavares and Pereira2018, Reference Malta, Paiva, Elisei, Tavares and Pereira2020) and included two or more genes (18S, 28S) and regions (ITS1, 5.8S, ITS2) of the rRNA gene array. Consequently, direct comparisons between our analysis and those in previous studies have their limitations. Nevertheless, there are several points of agreement between one or more of those previous analyses, and the results of this study are as follows: (1) R. longispicula and R. lophii place together in a common clade; (2) R. longispicula and R. lophii are part of a clade that also contains Hysterothylacium longilabrum Li, Liu & Zhang, 2012; (3) R. brasiliensis Moravec & Thatcher, Reference Moravec and Thatcher1997 and R. moraveci Pereira, Tavares, Scholz & Luque, 2015 are sister taxa, but Raphidascaroides nipponensis Yamamguti, 1941 (the type species of the genus) does not cluster with them – that is, Raphidascaroides as currently construed is not monophyletic; and (4) Sprentascaris is recovered as a monophyletic lineage. Our results did not find Ichthyascaris to be monophyletic as in Malta et al. (Reference Malta, Paiva, Elisei, Tavares and Pereira2018, Reference Malta, Paiva, Elisei, Tavares and Pereira2020). Because of the different number of taxa used in this study, we cannot address this discrepancy. Perhaps more exhaustive analyses across a broader range of taxa will be needed to conclusively evaluate the status of Ichthyascaris as a valid genus.
The ML analysis in this study places R. mundeswariensis n. sp. in a clade that includes members of the subgenus Ichthyascaris – namely, R. lophii and R. longispicula. However, neither R. mundeswariensis nor R. gigi (which belong to the same clade) have the defining feature of Icthyasacaris – namely, the ‘anteriorly united lateral alae’ (Moravec & Justine, Reference Moravec and Justine2020). Therefore, we provisionally place the species from A. bato in the genus Raphidascaris without any subgenus designation at this time, pending more exhaustive analyses of raphidascaridids in the future.
Raphidascaris mundeswariensis is the fifth species of Raphidascaris to be described from freshwater environments. Its host, A. bato, locally called ‘Chengo’ in West Bengal, is an oxydurcine gobiid (Gobiidae: Oxydurcinae). The fish host is also unique in representing a monotypic genus. Excluding species of the subgenera Sprentascaris and Ichthyascaris, only four other valid species of Raphidascaris have been described from freshwater hosts – namely, the type species, R. acus (Bloch, 1779) and three others: R. cyprini Wang, 1965, R. leiocassis Wang, 1965 and R. gigi. Raphidascaris acus is a widely distributed parasite of the northern pike, Esox lucius Linnaeus, 1758, in the Holarctic and circumboreal regions, and exceptionally of brown trout, Salmo trutta Linnaeus, 1758, in certain locations in Europe (Smith, Reference Smith1984; Moravec, Reference Moravec1994). Raphidascaris cyprini was reported from the freshwater bagrid catfish Tachysurus dumerili (Bleeker, 1864) and Cirrhina sp. in China (Wang, Reference Wang1965; Li et al., Reference Li, Gibson and Zhang2016). Finally, R. gigi is a parasite of another freshwater bagrid catfish Tachysurus nudiceps (Sauvage, 1883) (=Pelteobagrus nudiceps) and Masu salmon Oncorhynchus masou (Brevoort, 1856) in Japan (Moravec & Nagasawa, Reference Moravec and Nagasawa2002). There does not appear to be any common pattern in the biogeography of these five freshwater species of Raphidascaris, and it appears that they evolved by opportunistic colonization of disparate fish hosts in different regions.
Sood (Reference Sood2017) transferred as many as 20 species of Hysterothylacium Ward & Magath, 1917 reported from South Asia to Raphidascaris – namely, Hysterothylacium carutti Lakshmi & Rao, 1993, Hysterothylacium channai Lakshmi, 1995, Hysterothylacium elurensis Lakshmi & Lakshmi, 1995, Hysterothylacium fossilii Lakshmi, 1996, Hysterothylacium ganeshi Lakshmi & Sreeramulu, 2007, Hysterothylacium japonicum Lakshmi, 1996, Hysterothylacium karanensis Lakshmi, 2011, Hysterothylacium kiranii Lakshmi, 1993, Hysterothylacium krishnai Lakshmi, 1992, Hysterothylacium longicaecum Lakshmi & Rao, 1993, Hysterothylacium lysani Lakshmi & Sreeramulu, 2008, Hysterothylacium narayansis Lakshmi, 1997, Hysterothylacium nellorensis Lakshmi, 1996, Hysterothylacium neocornutum Lakshmi, Rao & Shyamasundari, 1992, Hysterothylacium poecilurai Lakshmi & Sreeramulu, 2005, Hysterothylacium pseudotumbili Lakshmi, Rao & Shyamasundari, 1991, Hysterothylacium punctati Lakshmi, 1995, Hysterothylacium ritai Lakshmi & Sreeramulu, 2006, Hysterothylacium shamimi Gupta & Begum, 2007 and Hysterothylacium vinodae Gupta & Begum, 2007. The proposed combination of Sood (Reference Sood2017) appears to be based on the old, and by now rejected, synonymy of Ward and Magath's Hysterothylacium with Raphidascaris by Hartwich (Reference Hartwich, Anderson, Chabaud and Willmott1974). All the aforementioned species of Hysterothylacium are well documented as having both a ventricular appendix and an intestinal caecum (see Sood, Reference Sood2017 for details), a typical feature of Hysterothylacium (Deardorff & Overstreet, Reference Deardorff and Overstreet1980; Soota, Reference Soota1983). Hence, by definition, they are not species of Raphidascaris.
Soota (Reference Soota1983) provided a more realistic account of nominal Raphidascaris species from India and tentatively listed two species of Raphidascaris – namely, R. acus and Raphidascaris panijii Khan & Yaseen, 1969 – from the subcontinent. The record of R. acus was based on two female specimens ‘doubtfully recorded’ (Soota, Reference Soota1983) by Soota & Chaturvedi (Reference Soota and Chaturvedi1971) from a marine/estuarine fish host, Clupea sp., at Porbandar, Gujarat, India. As noted above, R. acus is primarily a parasite of northern pike, E. lucius, in the Holarctic and circumboreal regions (Smith, Reference Smith1984; Moravec, Reference Moravec1994) and its presence in Clupea sp. from Indian waters is unlikely. Raphidascaris panijii was incompletely described by Khan & Yaseen (Reference Khan and Yaseen1969), based on a single male worm from Khulna, Bangladesh. Soota (Reference Soota1983) considered the illustration of the purportedly male worm by Khan & Yaseen (Reference Khan and Yaseen1969) to be ‘that of a juvenile female’ and called the record ‘purely tentative’. Smith (Reference Smith1984) also questioned the validity of this species.
In conclusion, the present study provides molecular and morphological evidence of the first credibly documented Raphidascaris species, R. mundeswariensis n. sp. from the Indian subcontinent. To our knowledge, this is not only the first adult helminth to be recorded from A. bato but also the first adult nematode to be reported in any species of mudskipper. It is likely that two previously reported Raphidascaris species from freshwater fishes in the region – namely, R. acus and R. gigi – are misidentifications of other species. Thus, future research on the Indian subcontinent must focus on using an integrative approach – that is, a combination of molecular and morphological data – to clarify the obscure species diversity, host ranges and the evolutionary history of these poorly studied ascaridoids in this region.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0022149X2100033X
Acknowledgements
The authors express their gratitude to two anonymous reviewers who provided valuable suggestions and comments, and to Homargha Das (USIC) of The University of Burdwan for technical support of SEM. A.C. acknowledges the use of the St Norbert College Electron Microscopy Suite and Hitachi TM 3030+ SEM, Henry Hong, formerly of the University of Toronto, for providing the SEM specimen processing protocol, and T.J. Fayton for running the Bayesian analysis on CIPRES.
Financial support
This study was partly based on a PhD thesis of the first author (B.K.P.) and was supported by the State Fund of West Bengal (B.K.P., grant number FC(SC.)/RS/SF/ZOO./2018-19/36), the University Grants Commission (A.A., grant number F-30-383/2017(3SR)) and the Department of Higher Education, Science and Technology and Biotechnology, Government of West Bengal (A.A., project number 279(Sanc.)/ST/P/S&T/2G-04/2017). A.C. acknowledges support from the Division of Natural Sciences, St Norbert College.
Conflicts of interest
None.
Ethical standards
The authors affirm that all procedures regarding this work concur with ethical standards of the relevant national and institutional guides on the care and use of fishes.





