Introduction
Prostaglandins (PGs) are central mediators of female reproductive events such as ovulation, fertilization, implantation, maintenance of pregnancy and parturition, acting by autocrine, paracrine and endocrine effects (Sales & Jabbour, Reference Sales and Jabbour2003). The biosynthesis of PGE2 is initiated with the release of arachidonic acid (AA) from membrane phospholipids through the action of phospholipases. The released AA is metabolised to PGH2 by either cyclooxygenase (COX)-1 or COX-2. PGH2 is then converted into PGE2 through the action of prostaglandin E synthases (PGESs). Two of these synthases are membrane-bound enzymes: mPGES-1 is a perinuclear protein induced by proinflammatory stimuli and predominantly coupled with COX-2; and mPGES-2 is mainly constitutively expressed as a Golgi membrane-associated protein and is functionally coupled with both COX-1 and COX-2 (Kudo & Murakami, Reference Kudo and Murakami2005). A third cytosolic PGES (cPGES) is constitutively expressed in a wide variety of cells and is functionally coupled to COX-1 to promote immediate PGE2 production (Kudo & Murakami, Reference Kudo and Murakami2005).
Although advances in bovine embryo in vitro production (IVP) technologies have occurred in recent years, the rate of early embryonic losses after transfer of IVP embryos is still high: between 56 and 81% of total pregnancy failures occur within 31 days after transfer of a single IVP embryo (for a total rate of pregnancy failures after IVP embryo transfer of 46–63%; Taverne et al., Reference Taverne, Breukelman, Perenyi, Dieleman, Vos, Jonker, de Ruigh and Van Wagtendonk-De Leeuw2002). More than 254,000 bovine IVP embryos at the morula and blastocyst stages were transferred worldwide in 2008 for genetic purposes (Thibier, Reference Thibier2009) but very little is actually known about the competence for post transfer development of these embryos (Hansen, Reference Hansen2006). Assessment of bovine embryo competence is usually made on morphological criteria but this is of relatively low predictive value. Numerous methods exist to evaluate the potential of embryo development and previous research allowed to identify some differences in gene expression related to developmental competence in bovine embryos (Wrenzycki et al., Reference Wrenzycki, Herrmann and Niemann2007). Efforts are still needed to find more accurate gene markers to identify the embryos best suited for transfer (Hansen, Reference Hansen2006).
Several data suggest that PGE2 plays an important role in the establishment of pregnancy in the cow. Bovine embryos collected between day 6 and day 10 after mating (morula to hatched blastocyst stages) from beef cattle metabolised AA primarily to PGE2 (Hwang et al., Reference Hwang, Pool, Rorie, Boudreau and Godke1988). After hatching from the zona pellucida, the bovine elongating blastocyst continues to secrete increasing amounts of PGE2 prior to, and throughout, the period of maternal recognition of pregnancy (days 10–18 post mating), in addition to PGF2α (after day 12) and PGI2 (after day 15; Hwang et al., Reference Hwang, Pool, Rorie, Boudreau and Godke1988; Wilson et al., Reference Wilson, Zalesky, Looney, Bondioli and Magness1992). Furthermore, for the PGs found in the bovine cyclic oviduct, PGE2 is the most abundant, its concentration peaking in the oviduct ipsilateral to the ovulation site during the postovulation phase (Wijayagunawardane et al., Reference Wijayagunawardane, Miyamoto, Cerbito, Acosta, Takagi and Sato1998). PGE2 originating from the bovine embryo could have paracrine and/or endocrine effects on both the oviduct and endometrium as several subtypes of the G protein-coupled PGE2 receptor (EP) are expressed in bovine oviductal cells during all stages of the oestrus cycle (EP2, EP3 and EP4; Gabler et al., Reference Gabler, Odau, Muller, Schon, Bondzio and Einspanier2008) and in uterine tissues during the first 50 days of pregnancy (EP2 and EP3; Arosh et al., Reference Arosh, Banu, Chapdelaine and Fortier2004a). PGE2 has been shown to play a key role in oviduct transport and passage into the uterus of equine embryos (Stout & Allen, Reference Stout and Allen2001; Weber et al., Reference Weber, Freeman, Vanderwall and Woods1991), in hatching from the zona pellucida in mouse and ovine blastocysts (Baskar et al., Reference Baskar, Torchiana, Biggers, Corey, Andersen and Subramanian1981; Sayre & Lewis, Reference Sayre and Lewis1993), and in blastocyst implantation in mice (Pakrasi & Jain, Reference Pakrasi and Jain2008). Pivotal roles of PGE2 in myometrial quiescence and luteal maintenance at the time of maternal recognition of pregnancy in cattle have also been proposed (Arosh et al., Reference Arosh, Banu, Kimmins, Chapdelaine, MacLaren and Fortier2004b). Recent data using bovine in vitro systems suggest that PGE2 is also involved in the regulation of oviduct contraction (Wijayagunawardane et al., Reference Wijayagunawardane, Miyamoto, Taquahashi, Gabler, Acosta, Nishimura, Killian and Sato2001) and in early embryo development (Marei et al., Reference Marei, Wathes and Fouladi-Nashta2009). Thus, embryonic PGE2 could be a major player in early embryo–maternal interactions leading to the origin and maintenance of pregnancy in cattle. The aim of this study was to investigate the mRNA expression of enzymes involved in the synthesis of PGE2 in IVP embryos produced from Holstein heifers.
Materials and methods
Animals
Holstein dairy heifers (n = 18), 16 to 20 months old, average live weight (LW) 367 ± 8.2 kg and located in the UNCEIA field station of Chateauvillain (Isere, France) were used for the study. Before the start of the experiment, estrous cycles were synchronized with subcutaneous 3 mg norgestomet ear implants (Crestar®, Intervet). On the day of implant insertion, heifers received an intramuscular (i.m.) injection of 3 mg of norgestomet and 3.8 mg of estradiol valerate (Crestar®). Two days before implant removal, heifers received 500 μg of cloprostenol IM (Estrumate®, Schering-Plough Vétérinaire). The animals came into heat 48 h after the removal of the implant and this time was used as the starting point for the ultrasound-guided transvaginal follicular aspiration (ovum pick-up).
Embryo in vitro production
Ovum pick-up (OPU) was performed as described by Guyader-Joly et al. (Reference Guyader-Joly, Ponchon, Thuard, Durand, Nibart, Marquant-Le Guienne and Humblot1997) twice a week over a period of 6 weeks on Mondays and Thursdays. Embryos used in this study came from each Monday OPU sessions i.e. six OPU sessions. All reagents originated from Sigma unless otherwise specified. Cumulus–oocyte complexes (COCs) with at least three layers of compact cumulus cells were incubated for 22–24 h in maturation medium (M199 medium supplemented with 10% fetal bovine serum (PanTM Biotech GmbH, 10 μg/ml FSH/LH (Stimufol®, Reprobiol), 1 μg/ml estradiol 17β and 5 ng/ml EGF) at 38.5°C in an humidified atmosphere of 5% CO2 in air. In vitro fertilization (IVF) was performed using frozen–thawed semen from a single ejaculate of a Holstein bull as described by Guyader-Joly et al. (Reference Guyader-Joly, Ponchon, Durand, Heyman, Renard and Menezo1999). The capacitation medium was a modified Tyrode's calcium-free medium (sp-TALP, pH 7.2) supplemented with 10 mM HEPES. Oocytes and capacitated spermatozoa were incubated for 18 h at 38.5°C in fertilization medium (Tyrode's solution (fert-TALP, pH 7.6) containing 10 μg/ml heparin-sodium salt, 20 μM penicillamine, 10 μM hypotaurine and 1 μM epinephrine). The day of IVF was defined as day 0. After removal of the cumulus cells, all zygotes from one heifer were cultured on a monolayer of Vero cells in 50-μl droplets of Upgraded B2 medium (Laboratoire CCD) covered with mineral oil at 38.5°C in a humidified atmosphere of 5% CO2 in air. At day 7, stage of embryo development were scored as either morula, early blastocyst, blastocyst or expanded blastocyst and their quality was graded according to classical morphological IETS criteria: grade 1 = excellent; grade 2 = good; grade 3 = medium; and grade 4 = poor quality (Stringfellow & Seidel, Reference Stringfellow and Seidel1988). Embryos were stored at –80°C until RNA extraction.
Embryo RNA extraction and cDNA preparation
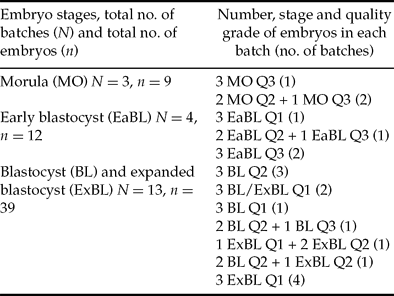
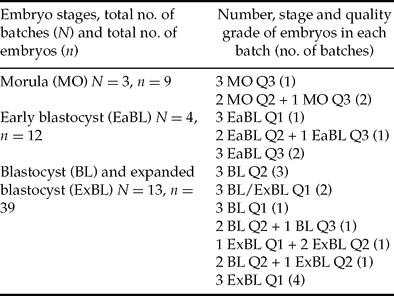
Only embryos of quality grades 1 to 3 were used in this study. RNA was extracted from batches of three day-7 IVP embryos at same or equivalent stages and qualities (see Table 1 for details). In order to compensate for experimental variation in RNA extraction and reverse transcription, all batches of embryos were spiked with 3 pg of exogenous luciferase RNA containing a poly(A) tail (Promega) before RNA extraction as previously described (Bui et al., Reference Bui, Evsikov, Khan, Archilla, Peynot, Henaut, Bourhis, Vignon, Renard and Duranthon2009). RNA extraction was performed using the RNeasy Mini Kit (Qiagen) and followed by a DNase-I treatment to remove genomic DNA (RNase-free DNase Set, Qiagen). Reverse transcription (RT) was performed using an oligo-dT and SuperScript III reverse transcriptase (Invitrogen). The reverse-transcribed cDNA was diluted five-fold (100 μl final volume) and stored at –20°C before use in real-time PCR.
Table 1 Batches of day 7 IVP embryos used in real-time PCR.
Real-time quantitative PCR (qPCR)
Primers sets used to amplify COX-1, COX-2, mPGES-1, mPGES-2 and cPGES were developed using known bovine sequences and designed using the Primer 3 web interface or adapted from the literature (see primers sequences in Table 2). In order to generate qPCR standards, RT-PCR products were amplified from 1 μg of bovine endometrium total RNA (for COX-2, mPGES-1, mPGES-2, cPGES), 1 μg of bovine corpus luteum total RNA (for COX-1) or 20 ng of luciferase RNA, purified using the Wizard PCR Preps DNA Purification System (Promega), quantified with a spectrophotometer and sequenced Beckman Coulter Genomics. Quantitative PCR reactions were performed in a 25-μl reaction mixture containing SYBR Green Master Mix (Applied Biosystems), 0.3 μM of each primer and 5 μl of each diluted RT reaction. A standard curve consisting of a ten-fold dilution series of the appropriate quantified amplicon, as well as no template controls, were included in each run. The reactions were performed on an ABI Prism 7000 (Applied Biosystems). For each sample, the PCR was carried out in triplicates: an initial denaturation step for 10 min at 95°C, then 45 cycles of 95°C denaturation for 15 s, 60°C annealing and extension for 60 s. Dissociation curves were performed after each PCR run to ensure that a single PCR product had been amplified. The AB Sequence Detection System software (7000 system SDS software, version 1.2.3, Applied Biosystems) was used to quantify and analyse the results. To account for potential variability between samples, the mean luciferase value obtained for each batch of embryos was used to correct the values obtained for each gene.
Table 2 Primers used in real-time PCR.

*Primers adapted from this reference to bovine sequences.
PCR specificity, sensitivity and efficiency
Specificity of the qRT-PCR products was validated by sequencing the PCR products and by analysis of the dissociation curve given by the SDS software (Applied Biosystems). The sensitivity and efficiency of the qPCR was evaluated using different starting amounts of purified standards. Values for r 2 and amplification efficiencies (E) were derived from the linear regression analysis of log(input cDNA) versus cycle number at threshold (Ct) plot. The slope created for each set of standards was used to determine E according to E = 10−slope – 1 where an E value of 1.0 would correspond to 100% cDNA replication at each cycle.
Statistical analysis
Normalisation of the results obtained for each gene within each batch of embryos was performed by calculating each as a ratio to the mean level of luciferase mRNA. Data are presented as means ± SEM. The F-test was used to examine the equality of variance and Student's t-test was used to compare mRNA ratios between embryo stages and qualities. Differences were considered statistically significant when p < 0.05.
Results
Real-time PCR validation
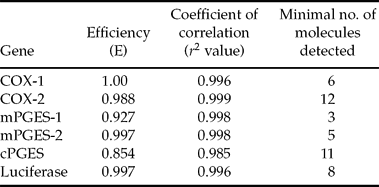
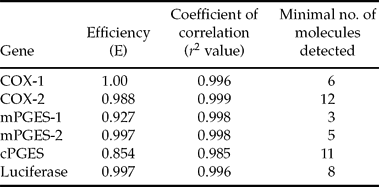
For each gene analysed, the sequence of the PCR product was 100% homologous with the corresponding bovine sequence on GenBank database. Furthermore, a single peak was obtained on the dissociation curves for each gene examined (data not shown). Within all qPCR assays, both efficiency and linearity were close to 1.0 and sensitivity was high (Table 3).
Table 3 Efficiency, linearity and sensibility of real-time PCR.
Expression of PGE2 synthases in embryos
The expression of enzymes involved in PGE2 synthesis was investigated by qRT-PCR in batches of three IVP embryos. Transcripts for COX-2 and mPGES-1 were detected in all batches of embryos. In contrast, transcripts for COX-1 and mPGES-2 were not detected in embryos whereas mRNAs for cPGES were at the limit of detection in eight batches of embryos (40%) and not detected in the others. Levels of COX-2 and mPGES-1 transcripts were significantly higher in blastocysts and expanded blastocysts (mRNA expression: 21.3 ± 4.0 and 44.0 ± 8.1 for COX-2 and mPGES-1, respectively; n = 13 batches of embryos) than in early blastocysts (5.3 ± 1.3 and 7.1 ± 1.3, p = 0.002 and p = 0.03 for COX-2 and mPGES-1, respectively; n = 4 batches of embryos) or than in morulae and early blastocysts taken together (5.7 ± 1.9 and 11.0 ± 3.0, p = 0.01 and p = 0.002 for COX-2 and mPGES-1, respectively; n = 7 batches of embryos; Fig. 1). Furthermore, levels of COX-2 and mPGES-1 were related to embryo quality based on morphological criteria. Embryos of excellent quality (grade 1) displayed higher mRNA expression of both COX-2 and mPGES-1 (25.2 ± 6.1 and 55.1 ± 11.4 for COX-2 and mPGES-1, respectively; n = 8 batches of embryos) than embryos of good (grade 2) and medium (grade 3) qualities (9.7 ± 2.2 and 16.2 ± 3.6, p = 0.04 and p = 0.01 for COX-2 and mPGES-1, respectively; n = 11 batches of embryos; Fig. 2).

Figure 1 Stage-dependant changes in (A) COX-2 and (B) mPGES-1 mRNA expression in batches of bovine IVP embryos at day 7. Each transcript level was normalised to luciferase expression. Data are means ± SEM of three replicates. Within each panel, bars with asterisks are different from others taken together (*p = 0.01 and **p = 0.002). MO = morulas (three batches of three embryos); EaBL = early blastocysts (four batches of three embryos); BL/ExBL = blastocysts and expanded blastocysts (13 batches of three embryos).

Figure 2 Quality grade-dependant changes in (A) COX-2 and (B) mPGES-1 mRNA expression in batches of bovine IVP embryos at day 7. Each transcript level was normalised to luciferase expression. Data are means ± SEM of three replicates. Within each panel, bars with different superscripts are different (*p = 0.04 and **p = 0.01). Grade 1 = excellent embryo morphology (eight batches of three embryos); Grade 2–3 = good and medium embryo morphologies (11 batches of three embryos).
Discussion
Our results suggest that IVP bovine embryos at the morula and blastocyst stages use exclusively the COX-2/mPGES-1 pathway for the biosynthesis of PGE2. The induction of both COX-2 and mPGES-1 expression has been observed in various systems in which PGE2 plays a critical role, such as inflammation, tissue repair or female reproduction (Kudo & Murakami, Reference Kudo and Murakami2005). The expression of COX-2 but not COX-1 has been previously reported in bovine preovulatory granulosa cells (Tsai et al., Reference Tsai, Wiltbank and Bodensteiner1996) and in ovine embryos collected from day 8 to day 17 after mating (Charpigny et al., Reference Charpigny, Reinaud, Tamby, Creminon and Guillomot1997). High expression of COX-2 mRNA with low expression of COX-1 has been also demonstrated in days 12 and 14 equine embryos (Aurich & Budik, Reference Aurich and Budik2005). In contrast, the expression of both COX-1 and COX-2 was showed at the transcript and protein levels in human in vitro-fertilised morulas and blastocysts (Wang et al., Reference Wang, Wen, Mooney, Behr and Polan2002), and at protein level in murine morulas and blastocysts collected in vivo (Tan et al., Reference Tan, Liu, Diao and Yang2005). However, in human embryos, COX-1 was mainly expressed during early stages of embryogenesis (zygote and 2-cell stages), whereas COX-2 was predominantly expressed later in 8-cell, morula and blastocyst stages (Wang et al., Reference Wang, Wen, Mooney, Behr and Polan2002). The predominant expression of mPGES-1 in bovine IVP embryos differs from the expression of all three PGESs (mPGES-1, -2 and cPGES) detected by immunostaining in mouse embryos at morula and blastocyst stages (Tan et al., Reference Tan, Liu, Diao and Yang2005). However, in the mouse, the signal for mPGES-1 immunostaining was at low level from the zygote to the 8-cell stages and became stronger at the morula and blastocyst stages, whereas cPGES signal decreased between the 4-cell stage and blastocyst stages (Tan et al., Reference Tan, Liu, Diao and Yang2005).
To our knowledge, the expression of all enzymes potentially involved in PGE2 synthesis has not been previously examined in bovine embryos. However, transcripts for COX-2 were reported in biopsies from IVP bovine blastocysts in a large-scale transcriptional analysis (El-Sayed et al., Reference El-Sayed, Hoelker, Rings, Salilew, Jennen, Tholen, Sirard, Schellander and Tesfaye2006). Furthermore, both mRNA and protein for COX-2 as well as for the three PGE synthases (mPGES-1, mPGES-2 and cPGES) were described in bovine immature and in vitro-matured cumulus-oocyte complexes (COCs) (Nuttinck et al., Reference Nuttinck, Guienne, Clement, Reinaud, Charpigny and Grimard2008). Interestingly, in this experiment, the expression of transcripts for mPGES-1 was 2- to 15-fold higher than those for both mPGES-2 and cPGES in bovine immature and matured COCs. Furthermore, during the maturation process, only the expression of both mPGES-1 and COX-2 significantly increased together with an increase in PGE2 release by COCs (Nuttinck et al., Reference Nuttinck, Guienne, Clement, Reinaud, Charpigny and Grimard2008), suggesting that the COX-2/mPGES-1 pathway was the main pathway responsible for PGE2 production during bovine oocyte maturation.
The expression levels of COX-2 and mPGES-1 mRNAs were higher in blastocysts and expanded blastocysts than in morula and early blastocyst stages. This finding suggests that the biosynthesis of PGE2 increases at the time of blastocoel expansion and may be involved in this process as well as in blastocyst hatching. Hwang et al. (Reference Hwang, Pool, Rorie, Boudreau and Godke1988) showed that bovine embryos collected in vivo from morula to hatched blastocyst stages metabolised AA primarily to PGE2 but, to date, the potential roles of PGE2 in the process of blastocyst expansion and blastocyst hatching have not been demonstrated. Furthermore, expression of both COX-2 and mPGES-1 were higher in grade 1 embryos than in embryos of lower quality. These later results suggest that high-quality IVP embryos, which have a greater developmental competence than lower grade embryos after intrauterine transfer (Hansen, Reference Hansen2006), have also a greater capacity to synthesize PGE2. A transcriptional analysis of biopsies from good-quality IVP bovine blastocysts showed that COX-2 mRNAs were higher expressed in embryos resulting in calf delivery compared with embryos that were resorbed after transfer to recipients (El-Sayed et al., Reference El-Sayed, Hoelker, Rings, Salilew, Jennen, Tholen, Sirard, Schellander and Tesfaye2006). Taken together, these results indicate that COX-2 and mPGES-1 could be good candidate gene markers of developmental competence in IVP bovine embryos.
In conclusion, our results suggest that bovine IVP morulae and blastocysts use the COX-2/mPGES-1 pathway for the biosynthesis of PGE2 and that this pathway could be involved in blastocyst expansion and developmental competence. Further studies are needed to determine the roles of PGE2 in bovine embryo physiology during early pregnancy.
Acknowledgements
This work has been supported by the French National Research Agency (GENANIMAL program) and APIS GENE. The authors wish to thank the staff of UNCEIA R&D Station in Chateauvillain for their technical assistance in collecting oocytes and Professor S. Chastant-Maillard (ENVA) for critical reading of the manuscript.