INTRODUCTION
Methionine adenosyltransferase (MAT; S-adenosyl-L-methionine synthetase, EC 2.5.1.6) catalyses the enzymatic condensation of ATP and L-methionine yielding AdoMet as the primary product, and pyro- and orthophosphate as by-products (Mato et al. Reference Mato, Alvarez, Ortiz and Pajares1997, Reference Mato, Corrales, Lu and Avila2002). AdoMet is an activated co-substrate of important crossroads of metabolic pathways. Firstly, AdoMet is the principal biological methyl donor in all known trans-methylation reactions in living organisms. Secondly, AdoMet is one of the precursors of polyamine biosynthesis. Thirdly, AdoMet plays an important role in the trans-sulphuration pathway to cysteine, one of the amino acids constituting the scavenging tripeptide glutathione. Finally, AdoMet is the main source of 5-deoxyadenosyl radicals, which are involved in DNA repair and in the biosynthesis of vitamins, coenzymes and antibiotics (Fontecave et al. Reference Fontecave, Atta and Mulliez2004; Reguera et al. Reference Reguera, Redondo, Perez-Pertejo and Balana-Fouce2007; Wang and Frey, Reference Wang and Frey2007).
The gene encoding MAT (MAT2 gene) in the trypanosomatid Leishmania infantum was cloned by our group, demonstrating that the genome of this parasite contains 2 identical copies of the gene separated by a long 3953-bp spacer region (Reguera et al. Reference Reguera, Balana-Fouce, Perez-Pertejo, Fernandez, Garcia-Estrada, Cubria, Ordonez and Ordonez2002). MAT2 genes are arranged in tandem and alternate with another gene, named LORIEN, which encodes a protein of unknown function with a SP-RING/Miz zinc-finger motif. The two copies of the LORIEN gene are located 3250-bp upstream of each MAT2 gene copy (García-Estrada et al. Reference Garcia-Estrada, Perez-Pertejo, Ordonez, Balana-Fouce and Reguera2007). Genomic studies have shown that these two genes are transcribed into large polycistronic nascent RNAs, which are post-transcriptionally processed into monocistronic mRNAs by sequential addition of a 39-nt spliced leader at the 5′-end and a poly(A) tail at the 3′-end (García-Estrada et al. Reference Garcia-Estrada, Reguera, Villa, Requena, Muller, Perez-Pertejo, Balana-Fouce and Ordonez2003).
MAT catalyses the only reaction that generates AdoMet in all organisms; therefore its role in cellular homeostasis is crucial. Drummelsmith and coworkers (Reference Drummelsmith, Girard, Trudel and Ouellette2004) carried out a proteomic analysis with a metothrexate (MTX)-resistant Leishmania strain observing that MAT abundance and AdoMet levels built up early during exposure to the drug, suggesting that increased MAT activity may facilitate the appearance of MTX resistance. These authors suggested a number of possible mechanisms that could lead to quick changes in MAT activity, including phosphorylation or post-translational modifications of the enzyme. Our laboratory previously developed a MAT-overexpressing L. donovani strain, which was able to synthesize 5-fold more MAT protein than the parental strain with the corresponding increase in AdoMet levels (Pérez-Pertejo et al. Reference Perez-Pertejo, Reguera, Ordonez and Balana-Fouce2006). However, despite the effective transcription rate of the exogenous gene throughout the culture time, MAT abundance and activity were effectively depressed to undetectable levels during the logarithmic phase of the culture. The rapid MAT depletion, in combination with an efficient efflux of the AdoMet excess to the culture medium, assures the parasite survival under a potential hypermethylation risk (Bacchi et al. Reference Bacchi, Garofalo, Ciminelli, Rattendi, Goldberg, McCann and Yarlett1993). All these results pointed to the existence of post-translational mechanisms able to adapt the intracellular AdoMet levels to those strictly required for cell survival, by means of rapid changes in MAT turnover.
It is well stated that the relative abundance of many proteins involved in a wide range of cellular processes is mediated by the ubiquitin (Ub)-proteasome system, which is in charge of proteolytic degradation in eukaryotic cells (Voges et al. Reference Voges, Zwickl and Baumeister1999). However, very few proteins to be degraded by this system have been reported in ancient eukaryotes so far (Van Hellemond and Mottram, Reference Van Hellemond and Mottram2000; de Diego et al. Reference de Diego, Katz, Marshall, Gutierrez, Manning, Nussenzweig and Gonzalez2001; Persson et al. Reference Persson, Jeppsson and Nasizadeh2003; Dubessay et al. Reference Dubessay, Blaineau, Bastien, Tasse, Van Dijk, Crobu and Pages2006), despite the identification of a functional Ub/26S proteasome mechanism in most protozoan parasites of medical interest (see Hunter, Reference Hunter2007 for a comprehensive review). In addition, genes encoding ubiquitin-conjugating enzymes have been annotated in different parasite Genome Projects (Carlton et al. Reference Carlton, Angiuoli, Suh, Kooij, Pertea, Silva, Ermolaeva, Allen, Selengut, Koo, Peterson, Pop, Kosack, Shumway, Bidwell, Shallom, van Aken, Riedmuller, Feldblyum, Cho, Quackenbush, Sedegah, Shoaibi, Cummings, Florens, Yates, Raine, Sinden, Harris, Cunningham, Preiser, Bergman, Vaidya, van Lin, Janse, Waters, Smith, White, Salzberg, Venter, Fraser, Hoffman, Gardner and Carucci2002; El-Sayed et al. Reference El-Sayed, Myler, Bartholomeu, Nilsson, Aggarwal, Tran, Ghedin, Worthey, Delcher, Blandin, Westenberger, Caler, Cerqueira, Branche, Haas, Anupama, Arner, Aslund, Attipoe, Bontempi, Bringaud, Burton, Cadag, Campbell, Carrington, Crabtree, Darban, da Silveira, de Jong, Edwards, Englund, Fazelina, Feldblyum, Ferella, Frasch, Gull, Horn, Hou, Huang, Kindlund, Klingbeil, Kluge, Koo, Lacerda, Levin, Lorenzi, Louie, Machado, McCulloch, McKenna, Mizuno, Mottram, Nelson, Ochaya, Osoegawa, Pai, Parsons, Pentony, Pettersson, Pop, Ramirez, Rinta, Robertson, Salzberg, Sanchez, Seyler, Sharma, Shetty, Simpson, Sisk, Tammi, Tarleton, Teixeira, Van Aken, Vogt, Ward, Wickstead, Wortman, White, Fraser, Stuart and Andersson2005; Berriman et al. Reference Berriman, Ghedin, Hertz-Fowler, Blandin, Renauld, Bartholomeu, Lennard, Caler, Hamlin, Haas, Böhme, Hannick, Aslett, Shallom, Marcello, Hou, Wickstead, Alsmark, Arrowsmith, Atkin, Barron, Bringaud, Brooks, Carrington, Cherevach, Chillingworth, Churcher, Clark, Corton, Cronin, Davies, Doggett, Djikeng, Feldblyum, Field, Fraser, Goodhead, Hance, Harper, Harris, Hauser, Hostetler, Ivens, Jagels, Johnson, Johnson, Jones, Kerhornou, Koo, Larke, Landfear, Larkin, Leech, Line, Lord, Macleod, Mooney, Moule, Martin, Morgan, Mungall, Norbertczak, Ormond, Pai, Peacock, Peterson, Quali, Rabbinowitsch, Rajandream, Reitter, Salzberg, Sanders, Schobel, Sharp, Simmonds, Simpson, Tallon, Turner, Tait, Tivey, Van Aken, Walker, Wanless, Wang, White, White, Whitehead, Woodward, Wortman, Adams, Embley, Gull, Ullu, Barry, Fairlamb, Opperdoes, Barrell, Donelson, Hall, Fraser, Melville and El-Sayed2005; Ivens et al. Reference Ivens, Peacock, Worthey, Murphy, Aggarwal, Berriman, Sisk, Rajandream, Adlem, Aert, Anupama, Apostolou, Attipoe, Bason, Bauser, Beck, Beverley, Bianchettin, Borzym, Bothe, Bruschi, Collins, Cadag, Ciarloni, Clayton, Coulson, Cronin, Cruz, Davies, De Gaudenzi, Dobson, Duesterhoeft, Fazelina, Fosker, Frasch, Fraser, Fuchs, Gabel, Goble, Goffeau, Harris, Hertz-Fowler, Hilbert, Horn, Huang, Klages, Knights, Kube, Larke, Litvin, Lord, Louie, Marra, Masuy, Matthews, Michaeli, Mottram, Muller-Auer, Munden, Nelson, Norbertczak, Oliver, O'neil, Pentony, Pohl, Price, Purnelle, Quail, Rabbinowitsch, Reinhardt, Rieger, Rinta, Robben, Robertson, Ruiz, Rutter, Saunders, Schafer, Schein, Schwartz, Seeger, Seyler, Sharp, Shin, Sivam, Squares, Squares, Tosato, Vogt, Volckaert, Wambutt, Warren, Wedler, Woodward, Zhou, Zimmermann, Smith, Blackwell, Stuart, Barrell and Myler2005), pointing to the presence of a universal eukaryotic protein degradation pathway (Bouzat et al. Reference Bouzat, McNeil, Robertson, Solter, Nixon, Beever, Gaskins, Olsen, Subramaniam, Sogin and Lewin2000).
The present paper reports on how leishmanial MAT is post-translationally controlled by regulatory proteins that can modify both abundance and activity. The use of specific proteasome inhibitors and Ub antibodies clearly show that this enzyme is proteolytically degraded by the leishmanial proteasome pathway to inactive peptides. In addition, in vitro and in vivo MAT phosphorylation suggests a possible link between those two processes in Leishmania.
MATERIALS AND METHODS
Reagents
DNA restriction enzymes were obtained from Boehringer Mannheim, Germany. Thermus aquaticus (Taq) polymerase was from Promega (Madison, WI, USA) and Pyrococcus furiosus (Pfu) polymerase was from Stratagene (La Jolla, CA, USA). Radioactive precursors, [2, 8-3H]-adenosine 5′-triphosphate, [γ 32P]-adenosine 5′-triphosphate and L-[35S]cysteine/methionine Pro-Mix™, were from Amersham (Amersham Biosciences, Little Chalfont, Bucks, UK). Proteasome inhibitors MG-132, MG-115, epoxomycin and lactacystin, were from Calbiochem® (EMD Biosciences, Inc., Germany). Rat brain protein kinase C (PKC) was from Roche (Mannheim, Germany). L. infantum H2A antiserum was kindly gifted by Dr Requena (Centro de Biologia Molecular “Severo Ochoa”, CSIC). The selection marker antibiotic G-418 and penicillin/streptomycin cocktail were purchased from Sigma Chemical Co. (St Louis. MO, USA). Two-dimensional electrophoresis reagents were from Bio-Rad (Hercules, CA, USA). All other chemicals and reagents were of the highest quality available.
Cell cultures
L. donovani promastigotes were propagated in a completely defined Medium 199, supplemented with 10% heat inactivated fetal calf serum (FCS) and penicillin/streptomycin cocktail (containing 50 U/ml penicillin, 50 μg/ml streptomycin). The parasite strain used in all experiments was L. donovani lysozyme 75 that was a gift from Dr Requena.
PX63NEO-MAT construction
Cloning of L. donovani MAT2 gene was performed as previously described (Reguera et al. Reference Reguera, Balana-Fouce, Perez-Pertejo, Fernandez, Garcia-Estrada, Cubria, Ordonez and Ordonez2002). L. donovani MAT2 gene cloned into pBluescriptSK(-), was excised with XmaI and NdeI, and cloned into a linearized PX63NEO vector for leishmanial over-expression. This plasmid has the gene that confers resistance to G418, which was used as selection marker.
Transfection of promastigotes
L. donovani promastigotes were transfected using the episomal construction pX63NEO-MAT or pX63NEO (‘mock’ transfections) as described elsewhere (Pérez-Pertejo et al. Reference Perez-Pertejo, Reguera, Ordonez and Balana-Fouce2006). Briefly, 40 million promastigotes were electroporated with 10 μg of plasmidic DNA, using a Gene Pulser II (Bio-Rad) at 2250 V/cm and 500 μF. After electroporation, cells were incubated at 26°C during 24 h in 10 ml of culture medium supplemented with 10% FCS. Selection was done adding 50 μg/ml of G418. Cultures were subpassaged after 4 days, always with the selection antibiotic. After 2 weeks, promastigotes were ready for different assays.
MAT Assay
MAT activity was assayed as described previously (Gil et al. Reference Gil, Pajares, Mato and Alvarez1997). The assay contained in 250 μl total volume, 5 mm L-methionine, 1 mm ATP (containing [2,8-3H]-adenosine 5′-triphosphate, 46 Ci/mmol, Amersham), 100 mm Tris-HCl, pH 8·0, 240 mm KCl, 12 mm MgCl2, and 10 mm dithiothreitol (DTT). The reaction was stopped with 4 ml of ice-cold water. Reaction mixtures were loaded onto AG 50W-X4 cationic-exchanger columns, washed twice with 10 ml of water and eluted with 4 ml of 3 m NH4OH. Samples, previously neutralized with 1 ml of glacial acetic acid, were measured in a scintillation counter using 10 ml of Optiphase-Hisafe 3 (Wallac) cocktail for aqueous mixtures. One unit of MAT activity is defined as the amount of enzyme that catalyses the formation of 1 nmol AdoMet per hour and per mg of protein. Protein was determined according to the method of Bradford (Reference Bradford1976).
SDS-PAGE and Western blotting
L. donovani promastigotes were harvested at different time-points during growth and washed twice with PBS. After sonication and centrifugation at 10 000 g for 20 min, the supernatant fraction was removed. Then, 15 μg of protein from each time-point were diluted in the loading buffer (60 mm Tris-HCl, pH 6·8, 2% SDS, 5% 2-mercaptoethanol, and 5% glycerol), heated in a boiling water bath for 5 min, and analysed by SDS-PAGE (10% acrylamide, 2·7% bisacrylamide) (Laemmli, Reference Laemmli1970). Proteins were electrotransferred onto PVDF membranes (Sigma) O/N at 25–30 V/cm, and the blots were blocked by incubation in 10 mm Tris-HCl, pH 7·5, 1 m NaCl, 0·5% Tween 20, 5% non-fat milk powder (w/v) for 1 h at room temperature. Primary antibody was added to this buffer, and the blot was incubated for 2 h. The blot was washed thoroughly in 10 mm Tris-HCl, pH 7·5, 1 m NaCl, 0·5% Tween 20 and then incubated with a secondary anti-rabbit antibody conjugated to horseradish peroxidase (Sigma). Antibodies were detected using 3,3′-diaminobenzidine as substrate (Bio-Rad).
Immunoprecipitation
For immunoprecipitation of leishmanial MAT, parasites (3×107 promastigotes) were harvested by centrifugation, washed with PBS and treated with 100 μl of lysis buffer containing 50 mm Tris-HCl (pH 7·5), 150 mm NaCl, 1% Triton X-100, and a protease inhibitor cocktail (1 mm PMSF, 8 μg/ml of leupeptin, 4 μg/ml of pepstatin and 4 μg/ml of aprotinin). The mixture was incubated at 4°C for 30 min with gentle shaking, and sonicated for 10 min. The lysates were microcentrifuged at 4°C for 15 min. Seventy-five μl of soluble proteins were incubated with 60 μg of a monoclonal anti-Ub antibody (Biomol International, UK) for 12 h at 4°C with gentle shaking. Fifteen μl of Protein A-agarose slurry (Sigma-Aldrich) were equilibrated in 50 μl of lysis buffer and added to the immunoprecipitated mixture. After incubation on an orbital rotator for 1 h at 4°C, the beads were collected by centrifugation and washed 3 times in 0·4 ml of buffer A (10 mm Tris-HCl, pH 8·0, 30 mm NaCl, and 2% Triton X-100), twice in 0·4 ml of buffer B (10 mm Tris-HCl, pH 8·0, 50 mm NaCl, and 0·1% Triton X-100) and once in 0·4 ml of buffer C (10 mm Tris-HCl, pH 8·0 and 0·05% Triton X-100). Finally, the beads were resuspended in 60 μl of Laemmli buffer. Immunoprecipitated proteins were resolved by SDS-PAGE on acrylamide gels.
Pulse-chase experiments
Sixty million promastigotes were harvested by centrifugation, washed in PBS and pre-incubated for 30 min in 400 μl of methionine-free Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FCS, to deplete cold methionine from the cells. Cells were washed once with PBS and then pulse-labelled for 7 h with 5 μCi/ml L-[35S]cysteine/methionine Pro-Mix™ (Amersham®), in pre-warmed methionine-free 10% FCS-DMEM. Pulse-labelling reaction was terminated by the addition of a 1000-fold excess of cold methionine. After 1 wash step with PBS, cells were subsequently chased for 12 h or 24 h in leishmanial culture medium, in the presence or absence of 5 μ m MG-132, as indicated. At the end-point of the chase, the chase medium was removed and the cells were washed once with PBS, and lysed in 0·1 ml of lysis buffer containing 50 mm Tris-HCl, pH 7·5, 150 mm NaCl, 1% Triton X-100 and protease-inhibitor cocktail (Boehringer). [35S]-Sulphur-labelled MAT was immunoprecipitated with rabbit anti-MAT antibodies, then separated by SDS-PAGE and detected by autoradiography.
In vitro MAT phosphorylation
Leishmanial MAT was phosphorylated in vitro by a commercial rat brain PKC (Roche) according to the following protocol. Twenty μg of purified MAT were labelled with 0·05 mU PKC in a phosphorylating buffer containing 20 mm Tris-HCl, pH 7·5, 1 mm CaCl2, 0·25 bovine serum albumin (BSA), 100 mg/ml phosphatidylserine, 20 mg/ml 1,2 dioleyl-sn-glycerol and 1 μCi [γ-32P]ATP. Reaction was carried out for 0, 2, 10 and 15 min at 30°C. Half of the samples were used to determine MAT activity and the remaining half were used to visualize the incorporated radioactivity. One ml of cold 100% TCA was added to the phosphorylation reaction for 10 min at 4°C and the acid-precipitable material was obtained by centrifugation at 12 000 g and washed twice. Pellets were resuspended in SDS/PAGE buffer and loaded onto 12% SDS/PAGE gels. After electrophoresis gels were dried and exposed for 12 h to an autoradiography film.
Two-dimensional gel electrophoresis
Standard two-dimensional gel electrophoresis (2-DE) experiments were performed using intracellular soluble proteins from either mock transfected or MAT-over-expressing leishmanial promastigotes. For the first dimension, aliquots of 500 μg were diluted to a final volume of 300 μl of rehydration buffer (8 m urea, 2% CHAPS, 40 mm DTT, 0·5% ampholytes 3–10). Samples were applied to the IPG-strip (17 cm pH 4–7 linear; Bio-Rad). Isoelectric focusing was carried out on a Protean® IEF cell (Bio-Rad) at 20°C and 50 μA/strip. Focusing parameters were described elsewhere (Cuervo et al. Reference Cuervo, de Jesús, Junqueira, Mendonca-Lima, Gonzalez, Betancourt, Grimaldi, Domoni, Fernández and Cupolillo2007). The strips were applied to the second dimension gel. Before the second dimension, proteins were reduced (10 mg/ml DTT) and alkylated (25 mg/ml iodoacetamide) in equilibration buffer (6 m urea, 2% SDS, 300 mm Tris–HCl, pH 8·8, 20% glycerol). Equilibrated IPG strips were separated across 12% SDS-PAGE gels using a vertical system (Bio-Rad) and standard Tris/glycine/SDS buffer. Gels were run at 40 mA/gel at 15°C. Five replicates were performed for each analysis.
Protein visualization and image analysis
Gels from 2-DE were stained with colloidal Coomassie Brilliant Blue G-250 (Tryoen-Tóth et al. Reference Tryoen-Toth, Richert, Sohm, Mine, Marsac, Van Dorsselaer, Leize and Florentz2003). Gel images were documented using a GS-800™ calibrated imaging densitometer (Bio-Rad) and image analysis was performed using PDQuest™ software (Bio-Rad). Comparison of 2-DE maps derived from 5 protein preparations, obtained from ‘mock’ and MAT-over-expressing promastigotes, was carried out. To determine experimental molecular mass for each single spot, 2-DE gels were calibrated using molecular mass markers (Bio-Rad).
Protein identification by mass spectrometry
In-gel digestions with trypsin and peptide recovery were performed as previously described (Drummelsmith et al. Reference Drummelsmith, Brochu, Girard, Messier and Ouellette2003). Peptide tandem mass spectra were obtained using a Q-TOF micro (Waters-Micromass) mass spectrometer. Mass measurements were transferred through the MS BioTools program (Bruker-Daltonics Billerica, MA, USA) as inputs to search the NCBInr database using the MASCOT software (http://www.matrixscience.com/). Peptide identification was confirmed by manual inspection of the spectrum.
RESULTS
MAT degradation is dependent on a protein or protein complex
In an attempt to determine MAT turnover, MAT-overexpressing L. donovani promastigotes were incubated in the presence of 10 μg/ml CHX. Culture aliquots were harvested at different time-points. For each time-point 1 aliquot was employed to determine MAT activity and another aliquot was used to assess MAT abundance in the same experiment. Fig. 1 shows that unexpectedly, the MAT from CHX-treated cultures turned over more slowly than the MAT from untreated cultures. This surprising result was corroborated through the analysis of MAT abundance by blotting 15 μg of leishmanial extracts to a PVDF membrane and hybridizing with a MAT polyclonal antibody. Fig. 1A shows that the band corresponding to a molecular mass of 47 000 Da (MAT monomer) disappeared almost completely 48 h after the beginning of the experiment, unlike MAT obtained from CHX-treated promastigotes (Fig. 1B), which remained visible for much longer. MAT activity was concordant with MAT turnover, showing only a 25% decrease at the end of the experiment in CHX-treated cells (Fig. 1C). MAT half-life was estimated either in the absence or presence of CHX, with values of 36 h and above 60 h, respectively. This experiment points to the presence of an auxiliary protein or protein complex, whose turnover is faster than MAT turnover and that may be involved in MAT degradation.

Fig. 1. Developmental expression of MAT along the growth curve of Leishmania donovani promastigotes. Culture was initiated at 1×106 promastigotes/ml and 24 h later, the culture at a density of 5×106 cells/ml was divided into two. One half was treated with 10 μg/ml CHX. The control culture grew normally, whereas the CHX-treated portion stopped its growth immediately. (A) Western analysis showing the MAT abundance of MAT-over-expressing L. donovani promastigotes after 24, 36, 48, 54 and 60 h. (B) Western analysis showing MAT protein abundance during the complete cell cycle (from 24 h to 60 h) of L. donovani promastigotes treated with 10 μg/ml CHX. Whole lysates harvested from L. donovani promastigotes (containing 15 μg of protein), were resolved by SDS-PAGE, blotted and incubated with a polyclonal-MAT antiserum. Loading controls made with a H2A antibody have been included below each blot. (C) MAT activity from L. donovani promastigotes, treated (○) or not (●) with 10 μg/ml CHX. Each spot represents the mean±s.d. of 3 representative experiments, using a paired t-test. ** P⩽0·001 regarding control; * P⩽0·01 regarding control cells.
MAT down-regulation is mediated by the proteasome
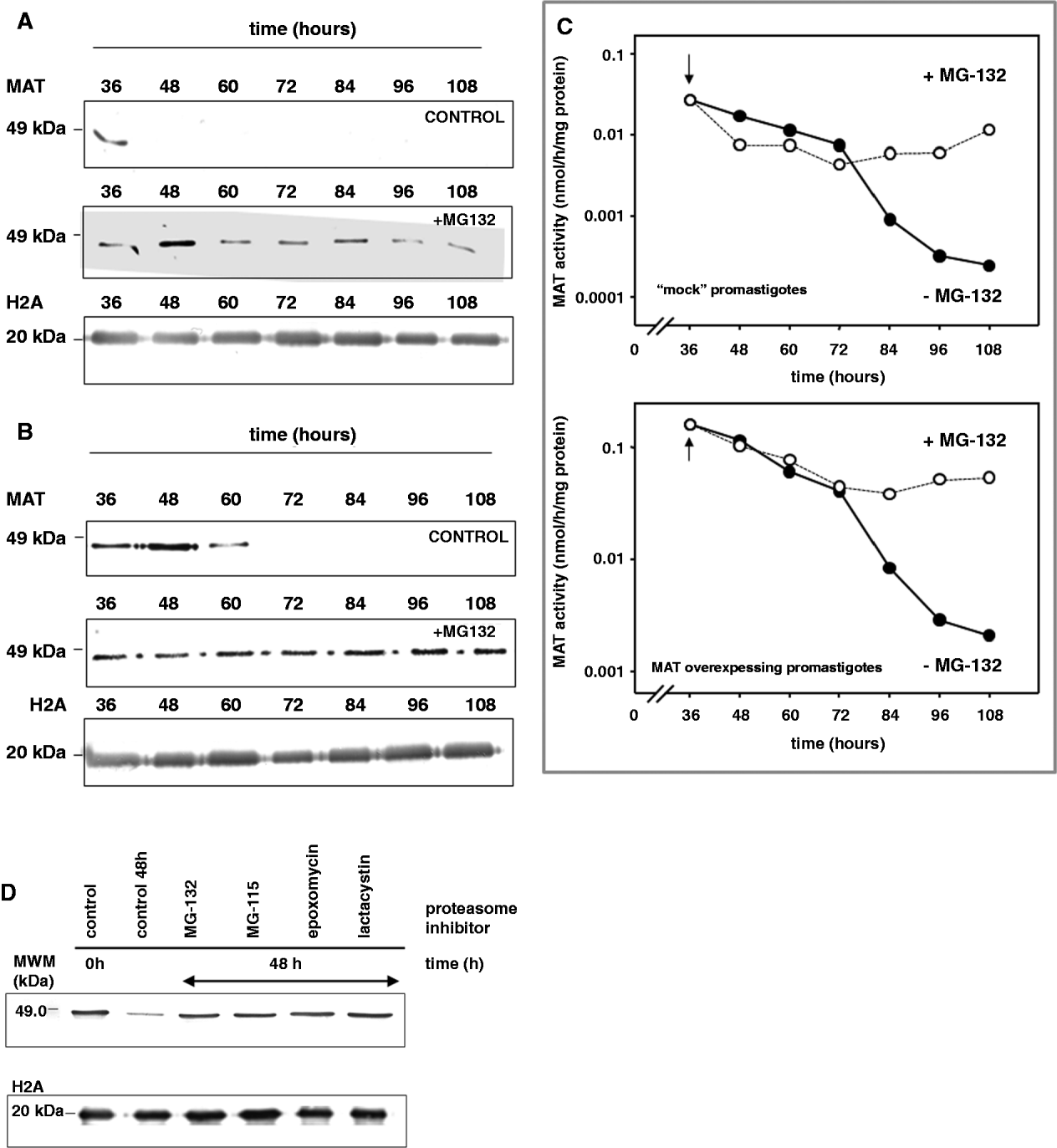
The possible involvement of leishmanial 26S proteasome in MAT down-regulation was studied using the specific inhibitor of proteasome MG 132. Mock-transfected and MAT-over-expressing L. donovani promastigotes were incubated in the presence of 5 μ m MG-132 and aliquots were harvested at different time-points. Fig. 2 shows the higher abundance of the 47 000-Da MAT immunoreactive band in the presence of MG 132, which provides evidence of the protective effect of the proteasomal inhibitor on MAT protein. In contrast, the MAT immunoreactive band disappears in the absence of MG 132. This phenomenon is common to the mock-transfected (Fig. 2A) and MAT-over-expressing (Fig. 2B) promastigotes pointing to a common MAT down-regulation mechanism in Leishmania parasites. With regard to MAT abundance, MAT activity was measured in MG 132-treated cultures with both ‘mock’ and MAT-over-expressing promastigotes (Fig. 2C). Unlike the cultures where the proteasome inhibitor was absent, those with MG 132 showed that the enzymatic activity was significantly retained. These differences between MG 132-treated and untreated cultures were more evident after 72 h.

Fig. 2. Effect of proteasome inhibitors on Leishmania donovani MAT stability. Cells were seeded at a density of 1 million/ml. After 24 h, the culture (at a density of 5×106 cells/ml) was divided and one half was treated with 5 μ m MG 132. The control culture grew normally, whereas the MG 132-treated culture stopped its growth. (A) Western analysis measuring MAT protein abundance of L. donovani promastigotes (‘mock-transfected’ strain) after 24, 48, 60, 72, 84, 96 and 108 h, in the absence and presence of 5 μ m MG 132. (B) Western analysis of MAT-over-expressing promastigotes after 24, 48, 60, 72, 84, 96 and 108 h in the absence and presence of 5 μ m MG 132. (C) MAT activity present in the lysates from ‘mock-transfected’ and MAT-over-expressing promastigotes untreated (●) and treated (○) with 5 μ m MG 132. The arrow represents the time-point for the addition of MG 132. (D) Western analysis of the lysates obtained from MAT-over-expressing promastigotes. Lane 1 represents the MAT-immunoreactive protein at time 0 (before the addition of the inhibitors), whereas lane 2 represents the same control (untreated cells) 48 h later. Lanes 3–6 correspond to leishmanial lysates treated for 48 h with 5 μ m MG 132 (lane 3), 5 μ m MG 115 (lane 4), 25 μ m lactacystin (lane 5) and 2·5 μ m epoxomycin (lane 6). Leishmanial histone 2A (H2A) was used as loading control.
In order to assess whether the prevention of MAT breakdown by MG 132 was an isolated event or, on the contrary, was due to the inhibition of the proteasome-pathway, other inhibitors of proteasomal proteolysis, namely MG-115 (5 μ m), lactacystin (25 μ m) and epoxomycin (2·5 μ m), were considered as potential protective agents of MAT degradation. Proteasome inhibitors were added to L. donovani promastigotes and incubated during 48 h. After this time, cells were harvested and washed. Total soluble proteins were loaded in a 10% SDS-PAGE gel, which was blotted onto a PVDF membrane subsequently incubated with MAT antibodies. Western analysis of the MAT-immunoprecipitated extracts shows that the immunoreactive band corresponding to MAT remained unchanged after 48 h (Fig. 2D) in the presence of the 4 proteasomal inhibitors, whereas the MAT-immunoreactive band disappeared almost completely in the control cultures lacking proteasome inhibitors.
Metabolic labelling of L. donovani promastigotes with L-[35S]cysteine/methionine in a free-methionine DMEM medium and immunoprecipitation with the polyclonal-MAT antiserum (pulse-chase experiments), shows a clear de novo synthesis of MAT in both ‘mock-transfected’ and the MAT-over-expressing promastigotes in the presence of MG-132 (Fig. 3). After 12 and 24 h, MAT-immunoreactive bands disappeared from the autoradiography film in those MG 132-untreated samples, thus suggesting an effective degradation of the de novo synthesized protein. However, when 5 μ m of MG-132 was added to the cultures, the MAT-immunoreactive bands remained visible along the experiment, indicating that protein degradation has been prevented. Altogether, these data indicate that MAT is effectively targeted by a proteasome-mediated system, which determines the abundance of this protein in leishmania promastigotes.

Fig. 3. Pulse-chase experiments of MAT stability in Leishmania donovani promastigotes; (A) ‘mock-transfected’ and (B) MAT-over-expressing L. donovani promastigotes were pulse-labelled for 7 h with L-[35S]-cysteine/methionine Pro-Mix™ (Amersham®), and chased for 0, 12 and 24 h in serum-free medium in the absence (−) or presence (+) of 5 μ m MG-132. The [35S]-labelled MAT was immunoprecipitated using anti-MAT antiserum and detected by autoradiography. The image shows a representative picture of 4 independent experiments.
Ubiquitination targets MAT for proteasome degradation
Immunoprecipitation experiments were carried out in order to test whether MAT degradation was mediated by a process involving protein ubiquitination. Wild-type promastigotes in the early exponential growth phase were treated with 5 μ m MG 132 and harvested at different time-points (0, 3, 6 and 12 h) by centrifugation, washed twice and lysed prior to immunoprecipitation with a commercial monoclonal anti-Ub antibody (Biomol International, UK). Immunoprecipitated proteins were loaded in a 10% polyacrylamide SDS-PAGE gel, which was blotted to a PVDF membrane subsequently incubated with MAT antibodies (Fig. 4). It is noteworthy that, in addition to the 47 000-Da MAT immunoreactive band, other bands corresponding to the mono- and di-ubiquitinated protein, were also detected. A clear smear from the ubiquitinated MAT-protein, was also visible, likely corresponding to poly-Ub tagged forms of MAT. At 12 h, control cultures showed very faint Ub-immunoreactive bands, unlike the MG-132-treated promastigotes, where the signal for the MAT protein was stronger. Those bands observed below the 47 000-Da MAT monomer may correspond to further ubiquitinated degradation peptides.

Fig. 4. MAT is ubiquitinated in vivo. Immunoprecipitation assays of Leishmania donovani promastigotes with Ub-antiserum. Wild-type cultures were harvested and lysed, and the MAT protein was immunoprecipitated using a specific monoclonal Ub-antiserum. Western blot analysis of the immunoprecipitated protein incubated with anti-MAT polyclonal antiserum. Lane C: immunoprecipitation assay using lysis buffer as sample. Lane 1: MWM. Lane 2: MAT at time 0. Lanes 3–5: time-dependence trend of ubiquitination at 3, 6 and 12 h after subpassage. Lanes 6–8: MAT degradation is prevented with 5 μ m MG-132 at the same harvesting times. The panel shows a representative picture of 3 independent experiments.
MAT phosphorylation for Ub degradation
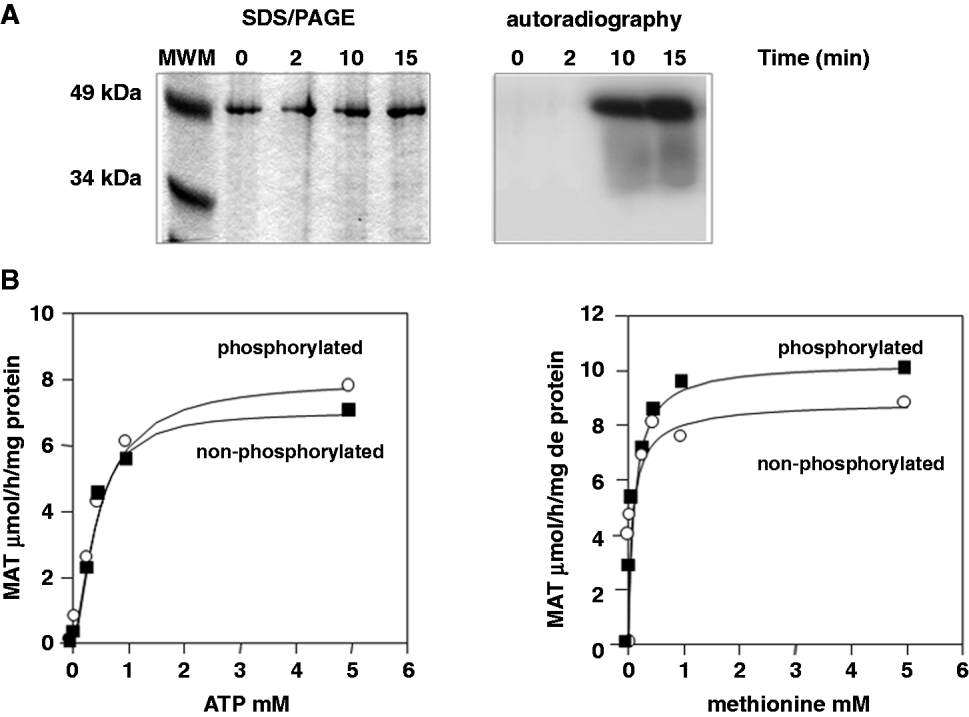
The connection of phosporylation and ubiquitination has been widely studied in mammals, where priming phosphorylation events may serve as a signal for further ubiquitination (Hunter, Reference Hunter2007). To test whether MAT has the potential to undergo phosphorylation events, in vitro experiments were conducted with the recombinant MAT. With this purpose, [γ-32P]-ATP and PKC were used following the protocol described in the Materials and Methods section. As shown in Fig. 5A, MAT is phosphorylated in vitro after 10 min of treatment with PKC. Studies were carried out to assess whether phosphorylation is able to affect MAT kinetics. As depicted in Fig. 5B, MAT activity remained constant irrespective of the phosphorylation events, which indicates that phosphorylation itself is not affecting the enzyme activity.

Fig. 5. Leishmanial MAT is phosphorylated in vitro, but its allosteric parameters are not modified. Three units of rat brain PKC were used to phosphorylate 5 μg of purified recombinant MAT from Leishmania donovani at different time-points. (A) Autoradiography of the SDS/PAGE showing in vitro MAT-phosphorylation. (B) MAT kinetics at variable concentrations of ATP and methionine saturation (left panel) and conversely, at variable methionine concentrations and ATP saturation (right panel) using a PKC-phosphorylated (○) and non-phosphorylated (●) recombinant MAT.
Since MAT was sensitive to phosphorylation in vitro, these results were confirmed in vivo using 2-DE gels. Proteins extracted from cultures of both the ‘mock-transfected’ and the MAT-over-expressing strain were analysed by 2-DE. Due to the high MAT amounts present in the over-expressing strain, identification of the spots containing the MAT protein was straightforward (spots 1A and 1B in Fig. 6). These spots were extracted from the gel, trypsinized and analysed by MS/MS as indicated in the Materials and Methods section. As shown in Table 1 (embedded in Fig. 6), the spots isolated from the gel corresponded unambiguously to the MAT protein. In addition, some of the peptides identified included phosphorylation events (see Table embedded in Fig. 6), thus confirming the results obtained in vitro. Since phosphorylation does not render changes on MAT allosteric behaviour (see Fig. 5B), phosphorylation may serve as priming signal for MAT degradation through the Ub pathway.

Fig. 6. MAT-over-expressing leishmanial promastigotes produce 2 isoforms of MAT with different phosphorylation patterns. Mock-transfected and MAT-over-expressing promastigotes were grown up to 24 h after subpassage. Cells were harvested and soluble extracts were obtained and analysed by 2-DE. The picture shows the enlarged region of 1 representative gel, which includes the MAT protein. The gel performed with MAT-over-expressing promastigotes shows 2 intense spots (1A and 1B indicated with arrows) clearly not visible in the mock-transfected cells. These spots were digested with trypsin and identified by MS/MS spectrometry, corresponding to the MAT protein. Some of the peptides contained phosphorylated amino acids (see embedded Table 1).
DISCUSSION
Several reports have demonstrated that intracellular AdoMet levels should be under the strict control of cellular processes in order to avoid undesirable hypermethylation status, which can be responsible for cell death (Reguera et al. Reference Reguera, Redondo, Perez-Pertejo and Balana-Fouce2007). An early report by Bacchi and coworkers (1993) showed that α-difluoromethylornithine (DFMO), the irreversible inhibitor of ornithine decarboxylase (ODC), could exert trypanocyde effect unbalancing the AdoMet/AdoHcy ratio in T. brucei trypomastigotes. Previous results obtained in our laboratory demonstrated that MAT-over-expressing L. donovani promastigotes maintained AdoMet under tolerable levels, which were compatible with cell survival (Pérez-Pertejo et al. Reference Perez-Pertejo, Reguera, Ordonez and Balana-Fouce2006). This strain was able to keep a high AdoMet efflux to the culture medium despite the high MAT activity (more than 5-fold compared to the control strain). In addition, MAT activity and MAT protein abundance were controlled by an unknown mechanism able to reduce their values near to those shown by the untransfected strain in a period of 48–72 h. Since trypanosomatids control gene expression mainly at the post-transcriptional level and MAT mRNA remained constant during the whole promastigote growing period, we postulate that mechanisms acting at the post-translational level are responsible for maintaining MAT activity at reasonable levels, which protects the cell from the hypermethylation pressure.
Early experiments were carried out in our laboratory in order to assess whether the leishmanial MAT was metabolically phosphorylated in an attempt to reproduce the post-translational modifications reported in mammalian cells (Pajares et al. Reference Pajares, Duran, Corrales and Mato1994). However, despite the fact that recombinant MAT protein is efficiently phosphorylated in vitro by a foreign PKC, no changes in allosterism were found, excluding this mechanism as a possible control system for MAT activity. An inter-relationship between post-translational processes in eukaryotic cells leading to protein activation and/or degradation has been recently suggested. The relationship between phosphorylation and ubiquitination has been shown in mammalian cells, but it still remains completely unknown in lower eukaryotes (Hunter, Reference Hunter2007). Phosphorylation acts as a positive crosstalk with ubiquitination by regulating the E3-ligase activity (a RING finger protein) of the final ubiquitination pathway or by creating phosphodegrons (short motifs that mediate phosphorylation-dependent recognition by E3-ligase) that drive the targeted protein to degradation. Conversely, protein kinase ubiquitination can lead to its proteasomal degradation. It is evident from the present results that MAT can be phosphorylated both in vitro (by PKC) and in vivo and that phosphorylation does not change the allosteric properties of the enzyme (like in mammalian cells). Therefore, we propose that MAT phosphorylation is a previous step to ubiquitination, promoting its proteasomal degradation, as it has been shown in immunoprecipitation experiments with the Ub antibody.
The major finding presented in the current work is that MAT enzyme is degraded by the leishmanial 26S proteasome. The proteasome inhibitors MG-132, MG-115, epoxomycin and lactacystin prevented MAT degradation after a lapse of 48 h, producing the accumulation of an active protein in both ‘mock-transfected’ and MAT-over-expressing promastigotes. Prevention of MAT degradation correlated well with the in vivo abolition of proteasome-proteolytic activity by the above-mentioned inhibitors. The 28-subunit cylinder-shaped structure composed of 4 stacked rings of 7 subunits each, which builds a 20S proteolytic complex called the catalytic core, has been described in most of the protozoan parasites of medical interest, namely Giardia lamblia (Emmerlich et al. Reference Emmerlich, Santarius, Bakker-Grunwald and Scholze1999), African (To and Wang Reference To and Wang1997; Yao et al. Reference Yao, Toth, Huang, Wong, Dias, Burlingame, Coffino and Wang1999) and American (González et al. Reference Gonzalez, Ramalho-Pinto, Frevert, Ghiso, Tomlinson, Scharfstein, Corey and Nussenzweig1996) trypanosomes, Leishmania spp. (Robertson, Reference Robertson1999; Silva-Jardim et al. Reference Silva-Jardim, Horta and Ramalho-Pinto2004) Entamoeba spp. (Hellberg et al. Reference Hellberg, Sommer and Bruchhaus1999) and the Apicomplexan P. falciparum (Li et al. Reference Li, Li, Mugthin and Ward2000) and Toxoplasma gondii (Paugam et al. Reference Paugam, Creuzet, Dupouy-Camet and Roisin2001).
Proteins to be degraded by the 26S proteasome complex are usually labelled by multiple Ub molecules, which are covalently attached to free amino groups on lysine side chains or to the N-terminal amino group of the targeted protein (Hicke et al. Reference Hicke, Schubert and Hill2005). Immunoprecipitation assays proved that MAT is mono- and di-ubiquitinated in Leishmania. To a lesser extent, poly-Ub tagged forms of MAT with a higher molecular mass seem to accumulate in MAT-immunoprecipitated promastigote extracts. As far as inhibition of the proteasome is concerned, low molecular weight degrading bands are missing from the Western blots incubated with MAT polyclonal antiserum, thus providing evidence of an effective inhibition of proteasome proteolytic activity.
Interestingly, the enzymes related to polyamine biosynthesis in mammalian cells are post-translationally regulated by proteasome degradation. In fact, both ornithine decarboxylase (ODC) and S-adenosylmethionine decarboxylase (AdoMetDC) are targeted by the 26S proteasome previous labelling with the specific antizyme (Pegg, Reference Pegg2006) or by ubiquitination (Yerlikaya and Stanley, Reference Yerlikaya and Stanley2004), respectively. In the case of protozoan parasites, Persson and co-workers (2003) reported divergences in turn-over control of ODC in T. brucei and Crithidia fasciculata. Despite their close phylogenetic resemblance, C. fasciculata ODC contains 2 PEST regions, conferring a much shorter half-life with respect to that of the African trypanosome. Heterologous transfection in a mammalian system with a plasmid containing the C. fasciculata ODC produces an unstable ODC with a slower turnover in the presence of the 26S proteasome inhibitor epoxomycin (Persson et al. Reference Persson, Jeppsson and Nasizadeh2003). This particular feature is in contrast to the mammalian ODC, which is degraded by the 26S proteasome in an Ub-independent way, where a specific ODC antizyme targets the protein into PEST regions before proteasomal degradation (Nasizadeh et al. Reference Nasizadeh, Jeppsson and Persson2003).
More recently, Dubessay and co-workers (Reference Dubessay, Blaineau, Bastien, Tasse, Van Dijk, Crobu and Pages2006) have described a nuclear kinesin (LmjKin13-1) from L. major, whose abundance seems to be dependent on proteasome degradation. Analysis of the amino acid sequence pointed to a double PEST-motif; the first one located on the N-terminal domain and the second one on the C-terminal end. Deletion analysis of these motifs using a GFP-engineered kinesin discarded the role of these signals in its turnover, pointing to the presence of alternative unknown labelling sequences in Leishmania.
In conclusion, the data presented in this work provide evidence of the down-regulation of a metabolic protein in both wild type and MAT-over-expressing L. donovani promastigotes. Taken together, these results confirm, for the first time that in unicellular parasites MAT is in vivo ubiquitinated and phosphorylated prior to proteasomal degradation.
ACKNOWLEDGEMENTS
We thank Dr José María Requena and his group (Centro de Biología Molecular Severo Ochoa, Universidad Autónoma de Madrid, Spain) for their help with molecular biology techniques. This study was supported by an AGL2009-11935 grant from MICINN and a PS09/00448 grant from Instituto de Salud Carlos III within the Network of Tropical Diseases Research (RICET).