Introduction
Arthropods are invertebrates that have developed the largest insect populations in nature during their long-term evolutionary process. Consequently, because of the important position and vital role of insects in nature, such as pollination and maintaining a balance between animals and plants, daily physiological and behavioral characteristics of insects should be intensively studied. Previous studies have detected the characteristics of circadian or seasonal periodic changes in a series of insect life activities and behaviors, such as growth, metamorphosis, reproduction, and avoidance of harm (Rosén, Reference Rosén2002; Shakeel et al., Reference Shakeel, He, Martin, Hanan and Wang2009; Seay and Thummel, Reference Seay and Thummel2011; Li et al., Reference Li, Yun, Yu and Li2018a). Therefore, it is believed that the biological clock is indispensable for ‘temperature compensation’ of fruit flies (Huang et al., Reference Huang, Curtin and Rosbash1995), diapause of silkworms (Xu et al., Reference Xu, Liang, Gan, Wang, Sima and Xu2011; Tobback et al., Reference Tobback, Boerjan, Vandersmissen and Huybrechts2012), and photoperiod activity of other insects (Goto and Matsumoto, Reference Goto, Matsumoto and Skinner2018). The circadian rhythm, a manifestation of the biological clock of insects, is a dynamic and endogenous oscillation that controls 24 h physiological and behavioral processes. Therefore, periodic activities of insects are the basis for unravelling the internal mechanisms of insect physiological behaviors and are subjects that require intensive study in integrated pest management.
Many studies have shown that there are obvious rhythmic phenomena in insect sexual behaviors, such as courtship and mating behaviors of Spodoptera exigua that occur during the first half of the scotophase (2–4 days after eclosion) (Luo et al., Reference Luo, Cao, Qian and Hu2003) and the sensitivity of male antennae (MA) to female sex pheromones (SPs), which is consistent with mating behavior in Spodoptera littoralis (Merlin et al., Reference Merlin, Lucas, Rochat, François, Maïbèche-Coisne and Jacquin-Joly2007). Some studies have confirmed that light, temperature, humidity, and chemical signals can regulate the expression of circadian clock genes and the biosynthesis, release, and perception of SPs in moths (Merlin et al., Reference Merlin, Lucas, Rochat, François, Maïbèche-Coisne and Jacquin-Joly2007; Tomioka and Matsumoto, Reference Tomioka and Matsumoto2015; Lu et al., Reference Lu, Huang, Liu, Wang, Chen, Xu, Deng and Ye2017; Chen et al., Reference Chen, Xu, Anantaprakorn, Rosing and Stanewsky2018). Before mating, female moths can biosynthesize and release SPs rhythmically through the SP gland (Groot, Reference Groot2014; Lu et al., Reference Lu, Huang, Liu, Wang, Chen, Xu, Deng and Ye2017). Age and mating could affect the courtship of male and female moths or other rhythmic behaviors (Groot, Reference Groot2014). These lines of evidence suggest that circadian clock genes may regulate SP communication in moths, but the detailed molecular mechanisms underlying the function of these genes remain unknown.
The insect rhythmic behavior is controlled by the circadian clock consisting of the negative transcriptional-translational feedback loop. It includes various circadian clock genes, such as Period (Per), Timeless (Tim), Cryptochrome (Cry), Cycle (Cyc), Clock (Clk), vrille (Vri), and Doubletime (Dbt), identified in numerous insects (Rothenfluh et al., Reference Rothenfluh, Abodeely and Young2000b; Tomioka and Matsumoto, Reference Tomioka and Matsumoto2015). The PAR domain protein gene (Pdp1ɛ) and the orange transcription factor gene (cwo) with the basic-helix-loop-helix (bHLH) domain might also be involved in the rhythmic expression process of circadian clock genes (Richier et al., Reference Richier, Michard-Vanhée, Lamouroux, Papin and Rouyer2008; Benito et al., Reference Benito, Hoxha, Lama, Lazareva, Ferveur, Hardin and Dauwalder2010). A master protein, PER, encoded by per binds to TIM (encoded by Tim), to form a heterodimer that performs a feedback function and coordinates the circadian and supernal rhythms of Drosophila melanogaster (Crosby et al., Reference Crosby, Hamnett, Putker, Hoyle, Reed, Karam, Maywood, Stangherlin, Chesham, Hayter, Rosenbrier-Ribeiro, Newham, Clevers, Bechtold and O'Neill2019) and the circadian rhythm activities of other moths, such as those involved in reproduction, mating, metamorphosis, and diapause (Beaver et al., Reference Beaver, Rush, Gvakharia and Giebultowicz2003; Tobback et al., Reference Tobback, Boerjan, Vandersmissen and Huybrechts2011). However, PER could also manifest heterotrophism by inhibiting dCLK/CYC-mediated transcription in Drosophila (Rothenfluh et al., Reference Rothenfluh, Young and Saez2000a). Additionally, some researchers have found that per gene is expressed rhythmically and non-rhythmically in different insects (Sauman and Reppert, Reference Sauman and Reppert1996; Lupien et al., Reference Lupien, Marshall, Leser, Pollack and Honegger2003) and has the same compensatory effect as Tim in expression regulation (Tobback et al., Reference Tobback, Boerjan, Vandersmissen and Huybrechts2012; Meuti et al., Reference Meuti, Stone, Ikeno and Denlinger2015). Studies have shown that Cry, a photoreceptor gene, might participate in the regulation or metabolism of TIM expression, which implies that environmental conditions could drive the diversification of the basic clock and its mechanisms (Chen et al., Reference Chen, Xu, Anantaprakorn, Rosing and Stanewsky2018), such as CRY1 with photoreceptors and mammalian-type CRY2 with repression (Ozturk, Reference Ozturk2016). CLOCK (CLK) and CYCLE (CYC), transcription factors consisting of a core feedback loop, might bind to each other and enter the nucleus to activate the transcription of period and timeless (Zhu et al., Reference Zhu, Sauman, Yuan, Casselman, Emery-Le, Emery and Reppert2008; Tokuoka et al., Reference Tokuoka, Itoh, Hori, Uryu, Danbara, Nose, Bando, Tanimura and Tomioka2017). However, there are structural differences between them in various insect species (Rubin et al., Reference Rubin, Shemesh, Cohen, Elgavish, Robertson and Bloch2006). Additionally, the transcriptional–translational loop is regulated by genes, such as Vrille (VRI) and Pdp1ε. Therefore, to fully understand the rhythmic sexual communication of insects, it is necessary to understand the causes of this rhythmic behavior in insects, namely the regulatory role of circadian clock genes.
Spodoptera litura (Lepidoptera: Noctuidae) is an important polyphagous pest. Its larvae feed on over 100 crops, and this pest is widely distributed throughout tropical and subtropical areas of Asia, including China (Dinesh-Kumar et al., Reference Dinesh-Kumar, Srimaan, Chellappandian, Vasantha-Srinivasan, Karthi, Thanigaivel, Ponsankar, Muthu-Pandian Chanthini, Shyam-Sundar, Annamalai, Kalaivani, Hunter and Senthil-Nathan2018; Liu et al., Reference Liu, Jia, Peng, Sheng, Tang, Xu, Han and Zhao2018; Wang et al., Reference Wang, Huang, Hao, Ran, Wu, Cui, Yang, Jiang and Yang2018; Li et al., Reference Li, Chen, Jin, Li, Wang and Shu2018b). Currently, the management of S. litura larvae mainly depends on chemical insecticides. Because of various side effects of chemical insecticides, including pollution of the environment and resistance phenomena of the target insects (Shad et al., Reference Shad, Sayyed, Fazal, Saleem and Zaka2012; Wang et al., Reference Wang, Huang, Hao, Ran, Wu, Cui, Yang, Jiang and Yang2018), there is an urgent need to develop alternative strategies for modern pest management to control insect populations. Although there have been many studies on SP communication in S. litura, some revealed that two desaturase genes (SlitDes5 and SlitDes11) (Xia et al., Reference Xia, Zhang, Ding, Wang and Lofstedt2019; Zhang et al., Reference Zhang, Zhang, Zhu, Zheng, Yan, Zhu, Xu, Zhang, He, Sun, Palli, Zhang and Dong2019), three pheromone-binding proteins (SlitPBP1-3) (Liu et al., Reference Liu, He and Dong2012; Liu et al., Reference Liu, Liu and Dong2013), and two odorant receptors (SlitOR6 and SlitOR13) (Zhang et al., Reference Zhang, Yan, Liu, Jacquin-Joly, Dong and Wang2015a) play important roles in SP biosynthesis and the perception processes; however, the specific regulatory mechanisms of the genes involved in this rhythmic behavior remain unknown. In the present study, sequences encoding putative candidate circadian clock genes in S. litura were identified through genome-wide transcriptome analysis. We performed a phylogenetic analysis of S. litura genes to establish the putative orthology of the different transcripts identified. Furthermore, we compared tissue specificity and temporal aspects of gene expression during development. The results could provide potential target genes to develop effective sexual communication disruptors to manage S. litura populations in the future.
Materials and methods
Insect rearing and tissue collection
Spodoptera litura larvae were reared at the College of Life Sciences, Huaibei Normal University, Huaibei, China, under laboratory conditions at a constant temperature of 28 ± 1°C, relative humidity of 60–70%, and a photoperiod of 14 h light: 10 h dark (LD). The eggs were incubated until hatching in an insect room, and various tissues were collected based on experimental purposes. To analyze the developmental expression levels of circadian clock genes, fresh eggs, 3- and 4-day-old larvae, pupae, and whole male and female adults were collected at 5 h after the beginning of the scotophase. To verify the tissue specificity of circadian clock genes expression, the following tissues were collected from virgin adults (3-day-old adults after eclosion and 5 h after the beginning of the scotophase) as follows: antennae (An), heads without antennae (He), thoraces (Th), abdomens (Ab), legs (Le), wings (Wi), hairpencils (Hp) of males, and SP glands of females (PG). To analyze the dynamic expression of circadian clock genes, SP glands and MA were collected from virgin female and male moths at 5–7 h after the beginning of the scotophase from different aged moths (0- to 5-day-old). Finally, to analyze the daily expression of circadian clock genes, we collected the SP glands and MA from virgin moths at different points from 2 to 3-day-old. The number of points was distributed according to the periods as 5 during the 14 h photophase, 6 during the 10 h scotophase, and 2 during the following 6 h photophase. At least three biological replicates were collected from each tissue and immediately stored in liquid nitrogen for later use.
RNA extraction and cDNA synthesis
According to experimental methods described previously (Zhang et al., Reference Zhang, Zhu, Fang, He, Wang, Chen, Sun, Ye, Deng and Li2015b), total RNA was extracted from tissue samples using the TRIzol® reagent (Tiangen Biotech, Beijing, China) following the manufacturer's instructions. Agarose electrophoresis and spectrophotometry were used to verify RNA quality. Next, the corresponding cDNA was directly synthesized using the PrimeScript™RT kit (TaKaRa, Dalian, China) according to the manufacturer's instructions and stored at −80°C until use.
Sequence and phylogenetic analysis
The genomeand transcriptomes of S. litura were downloaded from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/genome/?term=spodoptera + litura) (Cheng et al., Reference Cheng, Wu, Wu, Chilukuri, Huang, Yamamoto, Feng, Li, Chen, Guo, Liu, Li, Wang, Peng, Liu, Guo, Fu, Li, Liu, Chen, Tomar, Hilliou, Montagne, Jacquin-Joly, D'alencon, Seth, Bhatnagar, Jouraku, Shiotsuki, Kadono-Okuda, Promboon, Smagghe, Arunkumar, Kishino, Goldsmith, Feng, Xia and Mita2017). We first collected the circadian clock gene sequence of other insects from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and then, these genes were used as queries to screen the stand-alone genome of S. litura using NCBI-TBLASTN, with an E-value cut-off of e−5 for each query. The identified clock genes of S. litura were verified using NCBI BLASTX. The number and location of exons and introns of the circadian clock genes were generated using the Exon-Intron Graphic Marker (http://www.wormweb.org/exonintron). Open reading frames (ORFs) of all circadian clock genes were predicted using the ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Similarity searches were performed using the NCBI BLASTX network server (http://blast.ncbi.nlm.nih.gov/). The amino acid sequences of all clock genes of S. litura and other insects (table S1) were aligned using ClustalX 2.0, and phylogenetic neighbor-joining trees (1000 sampling analysis) were constructed using MEGA6.0 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
Measurement of mRNA levels
We used real-time quantitative PCR (qRT-PCR) to determine the specific expression patterns of clock genes and other genes related to SP communication in S. litura. First, the sample extraction mixture was used to detect the specificity of primers designed using Beacon Designer 7.9 (PREMIER Biosoft International, CA, USA) or referenced from other studies (Zhang et al., Reference Zhang, Zhu, Fang, He, Wang, Chen, Sun, Ye, Deng and Li2015b; Lin et al., Reference Lin, Qian, Bai, Zhang, Lu and Wan2017) (table S2). All PCRs were conducted in a 10 μl reaction volume containing 5 μl 2X SYBR Green PCR Master Mix (Vazyme Biotech, Nanjing, China), 0.25 μl of paired primers, 1 μl of cDNA, and increased to 10 μl with nuclease-free water. At least three technical replicates were performed for each biological sample. PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s. The final extension step was performed at 72°C for 10 min, and suitable aliquots were loaded onto a 2% agarose gel for visualization. Following the manufacturer's instructions, the melting curve was determined using the LightCycler®96 (Roche Diagnostics, Basel, Switzerland). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the elongation factor (EF) of S. litura were used as internal reference genes (Zhang et al., Reference Zhang, Zhu, Fang, He, Wang, Chen, Sun, Ye, Deng and Li2015b; Zhang et al., Reference Zhang, Li, He, Sun, Li, Fang, Ye, Deng and Zhu2016) to calculate the relative expression levels of each gene. The normalized expression of each gene was analyzed using Q-Gene (Muller et al., Reference Muller, Janovjak and Miserez2002; Simon, Reference Simon2003). Each biological sample had three technical replicates.
Statistical analysis
SPSS software (version 21.0; SPSS Inc., Chicago, IL, USA) was used for data processing and significance analysis (P < 0.05) using a one-way analysis of variance (ANOVA).
Results
Identification of candidate circadian clock genes
Based on the genome-wide annotation and comparative analysis in S. litura, we identified eight putative candidate circadian clock genes, including Per, Tim1, Tim2, Cry1, Cry2, Cyc, Clk, and Vri. The number of identified genes was more than Helicoverpa armigera, Bombyx mori, and Apis mellifera, and less than Danaus plexippus, D. melanogaster, Tribolium castaneum, and Acyrthosiphon pisum (fig. 1a). The corresponding sequence information of the eight circadian clock genes in S. litura is presented in table 1. According to the results of BLASTX match analysis, all clock genes were predicted to have full-length ORFs that encode 417–1284 amino acids and have a high level of identity among other noctuid pests (approximately 86–99%). The size and position of exons and introns were analyzed based on genome and cDNA sequences, and the results showed that SlitPer had the greatest number of exons and introns, whereas SlitCry2 had the least (fig. 1b). Next, six phylogenetic trees of insect circadian clock genes were constructed using various genes of S. litura and other insects (fig. 2). The results showed that each clock gene of S. litura formed a cluster on the same branch as the Lepidopteran species, and all branches were highly supported (bootstrap values greater than 60%).

Figure 1. Sequence information and structural characteristics of circadian clock genes in S. litura. (a) Distribution and comparison of circadian clock genes between S. litura and other insect species. (b) Intron and exon distribution of circadian clock genes in S. litura, scale bars represent a size of 500 bases.
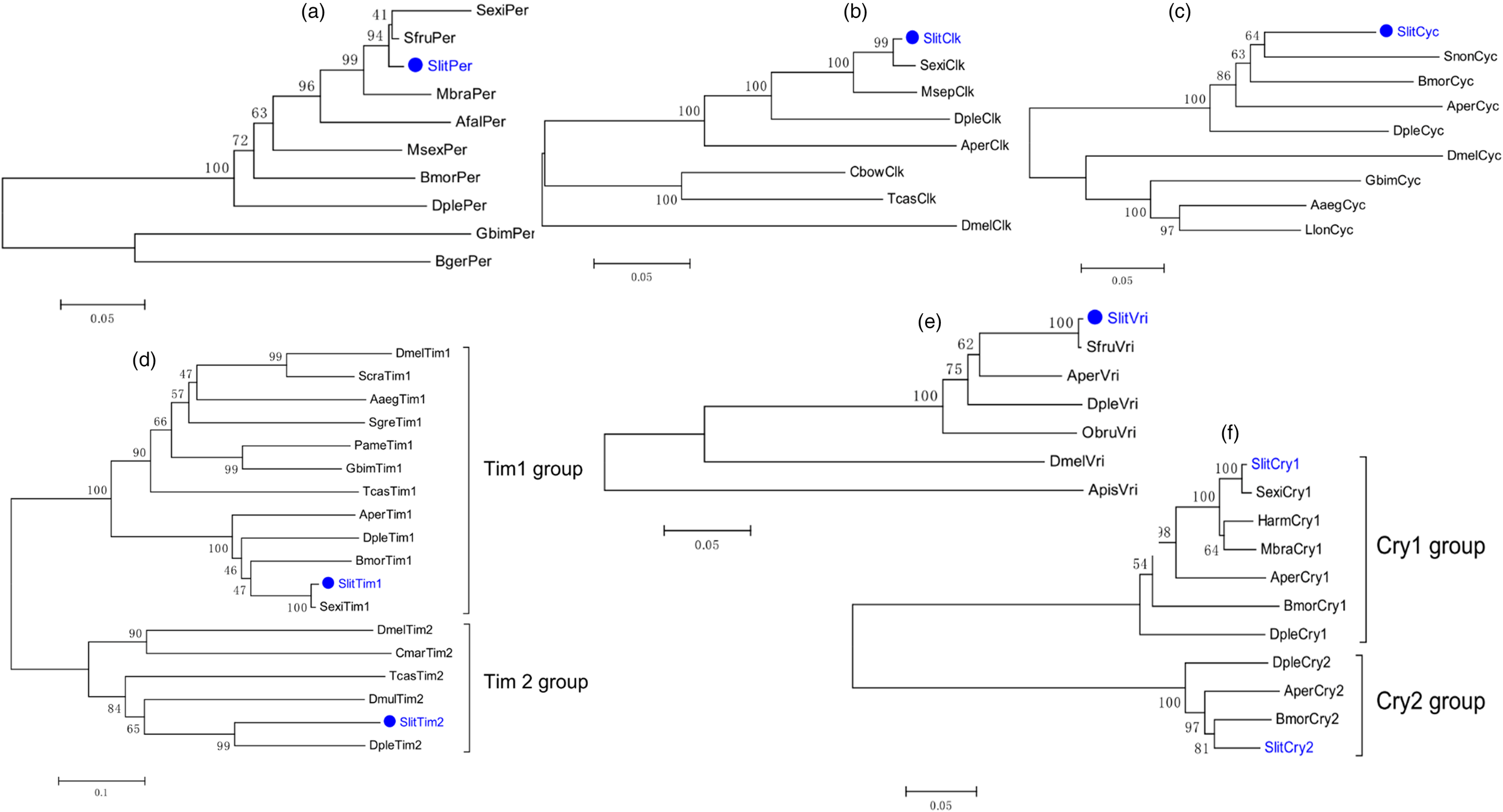
Figure 2. Phylogenetic trees of insect circadian clock genes. The S. litura translated genes are shown in blue. Other insects are as follows: Spodoptera exigua, Sexi; Spodoptera frugiperda, Sfru; Mamestra brassicae, Mbra; Anagrapha falcifera, Afal; Manduca sexta, Msex; Bombyx mori, Bmor; Danaus plexippus, Dple; Gryllus bimaculatus, Gbim; Blattella germanica, Bger; Antheraea pernyi, Aper; Mythimna separata, Msep; Colaphellus bowringi, Cbow; Tribolium castaneum, Tcas; Drosophila melanogaster, Dmel; Sesamia nonagrioides, Snon; Aedes aegypti, Aaeg; Lutzomyia longipalpis, Llon; Sarcophaga crassipalpis, Scra; Schistocerca gregaria, Sgre; Periplaneta Americana, Pame; Clunio marinus, Cmar; Diachasma muliebre, Dmul; Operophtera brumata, Obru; Acyrthosiphon pisum, Apis; Helicoverpa armigera, Harm. These trees were constructed using MEGA6.0 based on the alignment results of ClustalX2.0.
Table 1. The Blastx match of S. litura circadian clock genes.

H. armigera, Helicoverpa armigera; M. separata, Mythimna separata; S. exigua, Spodoptera exigua; S. frugiperda, Spodoptera frugiperda; ORF, open reading frame; aa, amino acid; MW, molecular weight; IEP, isoelectric point.
Developmental spectrum and tissue expression of clock genes in S. litura
To investigate the relationship between the expression of circadian clock genes relative to developmental stage, we analyzed the expression pattern of circadian clock genes at different developmental stages (from egg to adult) of S. litura using qRT-PCR (fig. 3). The results showed that all clock genes could be detected during each developmental stage of S. litura. Among these genes, the expression of SlitCry1 in eggs was significantly higher than that in the other developmental stages, and four genes (SlitPer, SlitCry2, SlitTim1, and SlitCyc) were significantly expressed in adults, three genes (SlitCry2, SlitTim2, and SlitCyc) were differentially expressed between the sexes in adults. Interestingly, we found that some genes in the same family showed different expression characteristics, such as SlitCry1, SlitCry2, SlitTim1, and SlitTim2. We further analyzed the expression levels of all clock genes in different tissues of unmated adults. The results showed that all the genes displayed significant sex differences (fig. 4). There was one highly expressed gene in the female antennae (SlitPer), heads (SlitCry2), and abdomens (SlitTim2), and two in the wings (SlitCyc and SlitVri), whereas there was no significant difference in expression in different tissues of SlitCry1. In contrast to females, five genes (SlitPer, SlitCry2, SlitTim2, SlitCyc, and SlitVri) in males exhibited significantly higher expression in the thoraxes and two genes (SlitCry1 and SlitTim1) were highly expressed in the MA and hair-pencils.
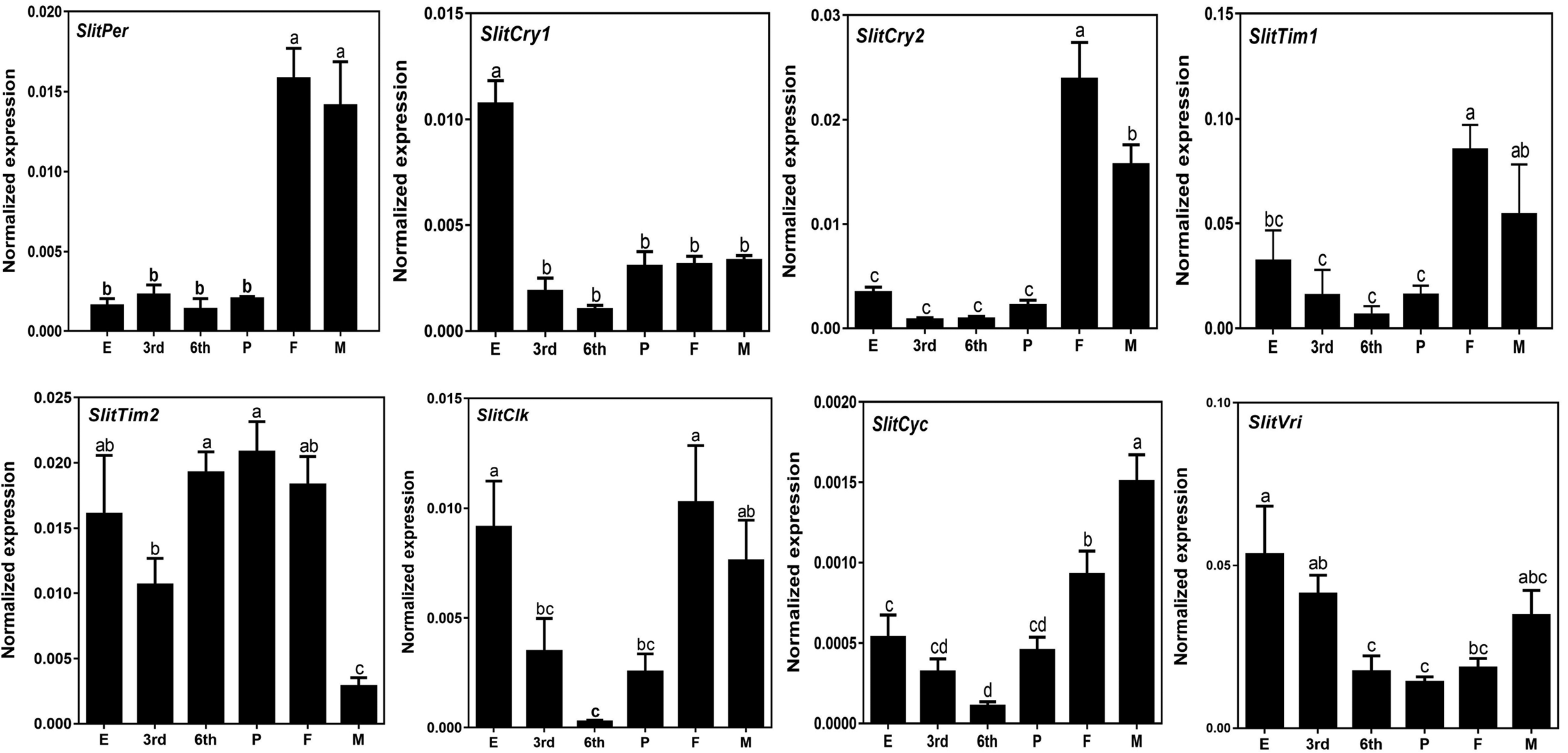
Figure 3. The expression analysis of circadian clock genes at different developmental stages of S. litura or others. The different lowercase letters mean significance between samples (P < 0.05, ANOVA, LSD). E, egg; 3rd, third larva; 6th, sixth larva; P, pupa; F, female; M, male. Horizontal axis: the different developmental stages of S. litura; vertical axis: the normalized expression of target gene was calculated by Q-gene software.
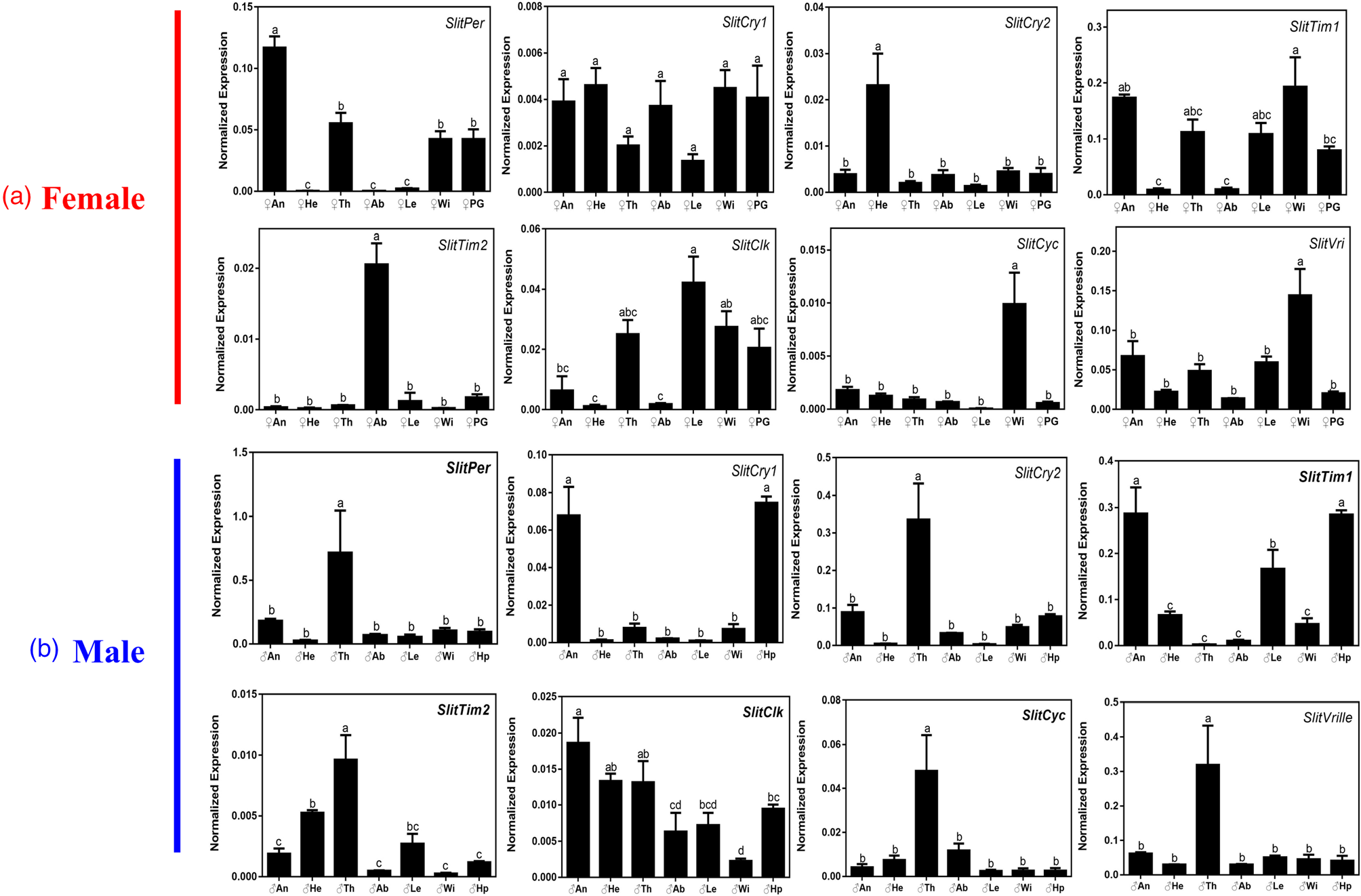
Figure 4. Tissue expression of circadian clock genes in S. litura. The different lowercase letters mean significance between tissues (P < 0.05, ANOVA, LSD). An, antenna; He, head without antenna; Th, thorax; Ab, abdomen; Le, leg; Wi, wing; Hp, hairpencil; PG, sex pheromone gland. (A) Female tissues, (B) Male tissues. Horizontal axis: the different tissue of adults S. litura; vertical axis: the normalized expression of target gene was calculated by Q-gene software.
Expression profiles of genes related to sex pheromone communication or circadian clock in adult S. litura
To determine whether there was a relationship between the dynamic distribution of SP communication-related genes and circadian clock genes in S. litura, we analyzed the dynamic expression of these genes using qRT-PCR. Based on previous studies (Liu et al., Reference Liu, Liu and Dong2013; Zhang et al., Reference Zhang, Yan, Liu, Jacquin-Joly, Dong and Wang2015a; Xia et al., Reference Xia, Zhang, Ding, Wang and Lofstedt2019; Zhang et al., Reference Zhang, Zhang, Zhu, Zheng, Yan, Zhu, Xu, Zhang, He, Sun, Palli, Zhang and Dong2019), some SP communication-related genes with confirmed functions were selected for the analysis, including SP biosynthesis-related genes (SlitDes5 and SlitDes11) in the female SP gland and SP perception-related genes (SlitOR6, SlitOR11, SlitOR13, SlitOR16, and SlitPBP1-3) in the MA. The results showed that the expression of SlitDes5 and SlitDes11 reached the highest level in 3-day-old PGs of S. litura (fig. S1), whereas other SP perception-related genes reached a peak in 4- or 5-day-old MA and six genes (SlitOR6, SlitOR11, SlitOR13, SlitOR16, SlitPBP2, and SlitPBP3) exhibited a rapid increase after 3- or 4-day-old. Regarding the circadian clock genes of S. litura, the qRT-PCR results showed that in female PGs, SlitPer and SlitCry2 had a similar dynamic expression pattern as that of SlitDes5 and SlitDes11 (fig. 5). However, in MA, all clock genes reached their peak levels at 2 or 3 days after eclosion, showing different expression patterns from those of SP perception-related genes (fig. 5).
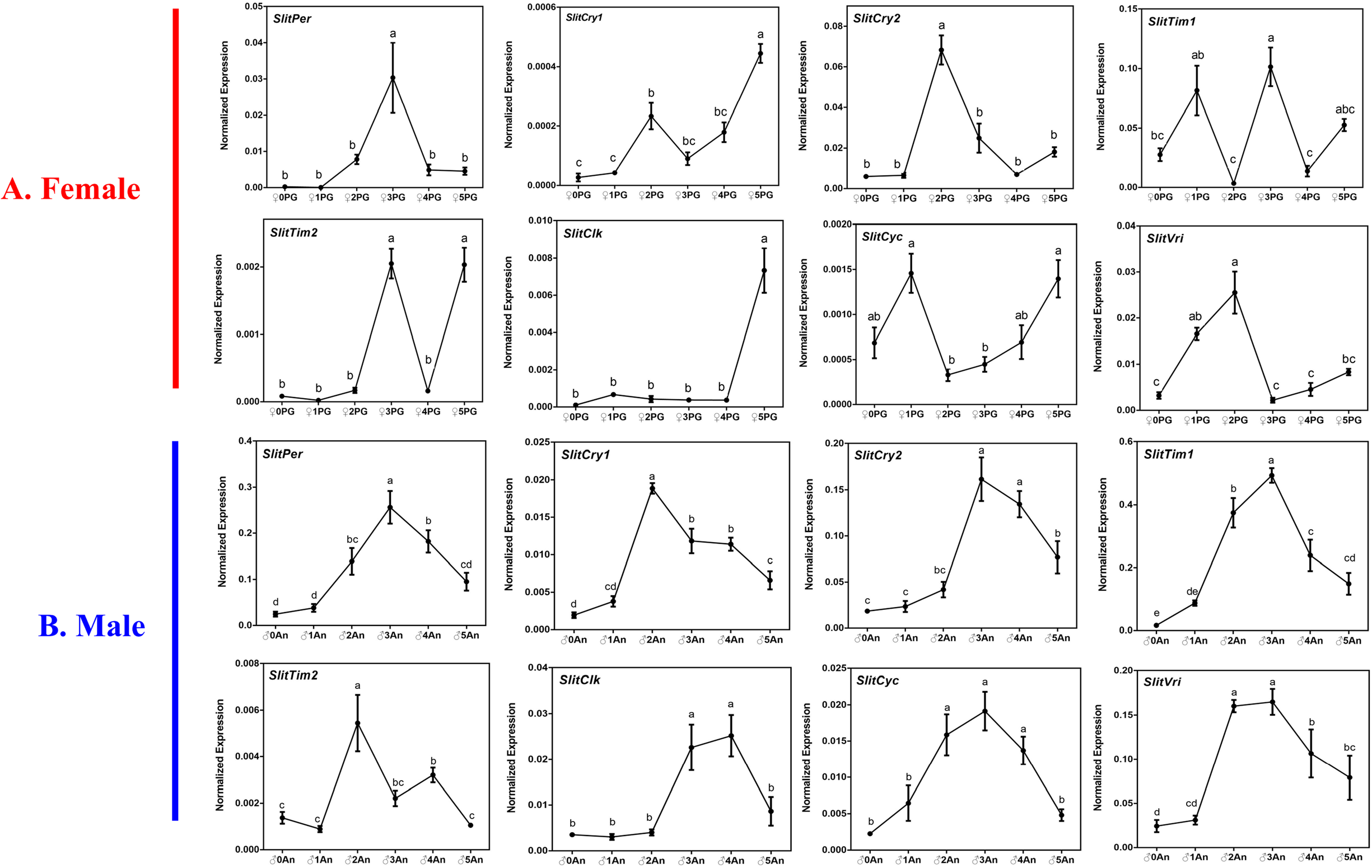
Figure 5. Dynamic expression of circadian clock genes in adult female sex pheromone gland (PG) and male antennae (MA) of S. litura. The different lowercase letters mean significance between tissues (P < 0.05, ANOVA, LSD). ♀, Female adult; ♂, male adult; PG, sex pheromone gland; An, antenna. Horizontal axis: the PG or An from virgin female or male moths at 5–7 h of the scotophase at different ages (0- to 5-day-old); vertical axis: the normalized expression of target gene was calculated by Q-gene software.
Daily expression profiles of genes related to sex pheromone communication or circadian clock in S. litura
Similar to other nocturnal moths, SP-mediated sexual communication of S. litura occurred mainly during the scotophase in 2- to 4-day-old adults (Sun et al., Reference Sun, Hu and Du2002). To explore whether the expression of SP communication-related and clock genes was related to the alternation of the photophase and scotophase, we analyzed daily expression of genes with samples collected during 14 h photophase (5 points), 10 h scotophase (6 points), and the ensuing 6 h photophase (2 points).
The qRT-PCR results showed that SlitDes5 and SlitDes11 in female PGs reached their expression peaks 6 and 2 h after the onset of scotophase, respectively (fig. S2). In MA, three SP perception-related genes (SlitOR6, SlitOR13, and SlitPBP1) peaked at the first point of the photoperiod, decreased gradually, peaked in the second hour of scotophase, and then decreased rapidly. However, SlitPBP2 reached its expression peak during the first hour of scotophase, and the expression peak for other SP perception-related genes (SlitOR11, SlitOR16, and SlitPBP3) did not occur during the scotophase; rather, a peak was reached during the photoperiod. The rise and decrease of the peak were not as rapid as that of other genes, suggesting the expression trend of the three genes in the entire LD cycle was different from that of the other genes (fig. 5).
We then measured the daily expression of all the S. litura clock genes in PGs and MA. Among all clock genes, SlitPer had the most similar daily expression pattern to SlitDes5 and SlitDes11 in PGs of S. litura. In the female PGs, the expression characteristics of SlitPer were that peak expression occurred at the sixth hour of the scotophase and the expression level increased rapidly after entering the scotophase and then decreased rapidly at the end of the scotophase (fig. 6). Although the expression peak of SlitCry2 appeared once in the photophase and once in the scotophase, the expression of SlitCry2 rapidly increased and then decreased after entering the scotophase, which was similar to the expression patterns of SlitDes5 and SlitDes11 in the scotophase (fig. S2). In the MA of S. litura, the qRT-PCR results showed that four clock genes (SlitPer, SlitTim2, SlitClk, and SlitCry2) and three SP perception-related genes (SlitOR6, SlitOR13, and SlitPBP1) had similar expression features; they decreased from the peak value in the first photoperiod and then reached the peak value at 14:00 after entering the scotophase (figs 7 and S2). There was no significant difference between the peak values in photoperiod and scotophase in the four clock genes. However, the peaks of SlitTim1 and SlitVri only appeared in the scotophase (14:00), but not in the photophase, and the peaks of SlitCyc and SlitCry1 appeared only in the photophase (1:00) (fig. 7).

Figure 6. Daily expression of circadian clock genes in adult female sex pheromone gland (PG) of S. litura. The different lowercase letters mean significance between tissues (P < 0.05, ANOVA, LSD). P, photophase; S, scotophase. Horizontal axis: the different points of samples were collected; vertical axis: the normalized expression of target gene was calculated by Q-gene software.
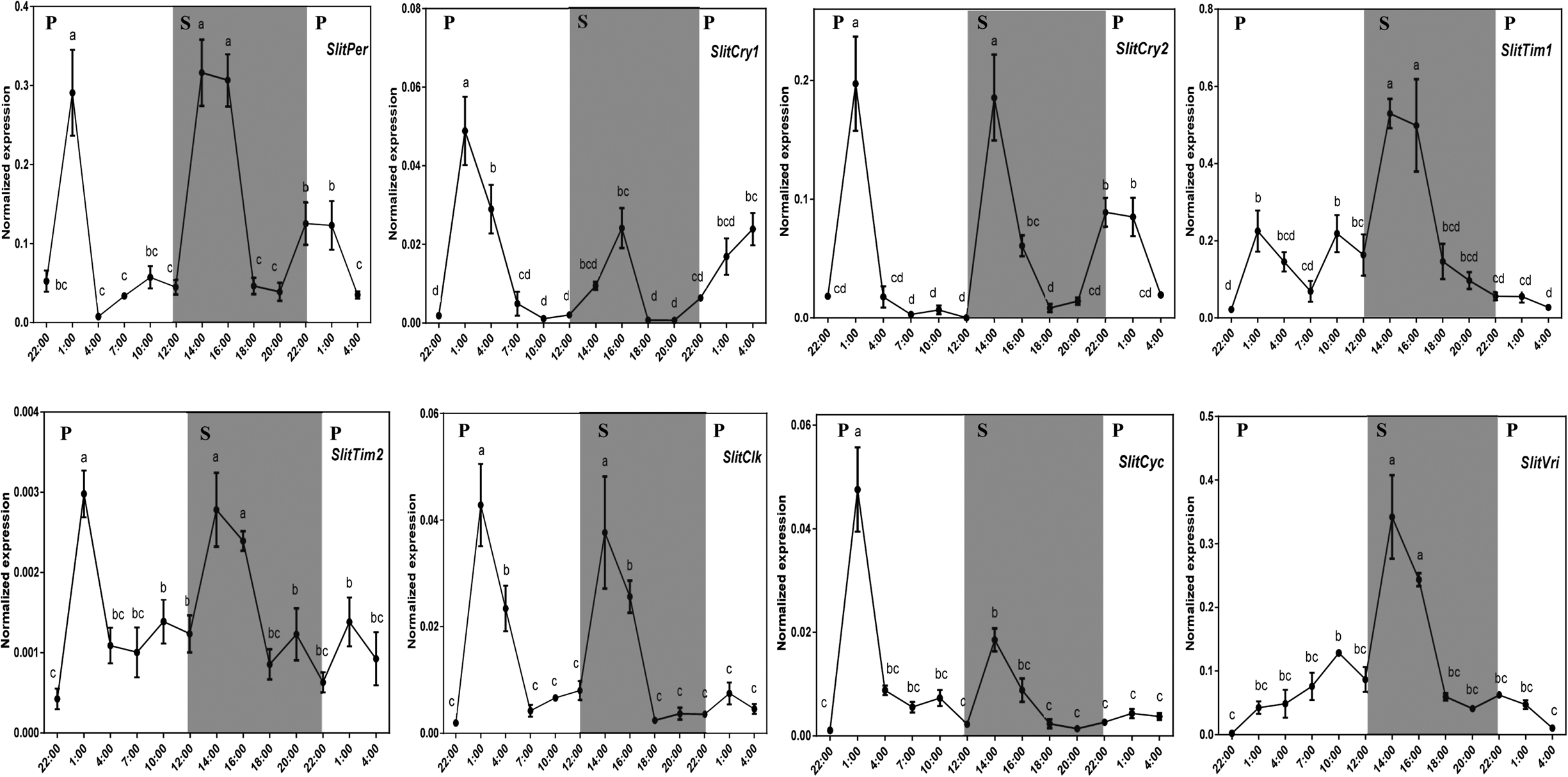
Figure 7. Daily expression of circadian clock genes in adult male antennae (MA) of S. litura. The different lowercase letters mean significance between tissues (P < 0.05, ANOVA, LSD). P, photophase; S, scotophase. Horizontal axis: the different points of samples were collected; vertical axis: the normalized expression of target gene was calculated by Q-gene software.
Discussion
To determine the possible involvement of the circadian clock machinery of sexual communication based on the SPs in S. litura, it is necessary first to understand the classification, molecular structure, and expression characteristics of circadian clock genes in key sexual communication tissues (PG and MA). In the present study, we obtained eight candidate circadian clock genes in S. litura based on genomic data (Cheng et al., Reference Cheng, Wu, Wu, Chilukuri, Huang, Yamamoto, Feng, Li, Chen, Guo, Liu, Li, Wang, Peng, Liu, Guo, Fu, Li, Liu, Chen, Tomar, Hilliou, Montagne, Jacquin-Joly, D'alencon, Seth, Bhatnagar, Jouraku, Shiotsuki, Kadono-Okuda, Promboon, Smagghe, Arunkumar, Kishino, Goldsmith, Feng, Xia and Mita2017), including SlitPer, SlitTim1, SlitTim2, SlitCry1, SlitCry2, SlitCyc, SlitClk, and SlitVri. Comparison with other Lepidopteran species whose genome sizes are presently known (Tomioka and Matsumoto, Reference Tomioka and Matsumoto2015) showed that the number of clock genes in S. litura is higher than that of H. armigera (7 clock genes) and B. mori (7 clock genes), but less than D. plexippus (9 clock genes). The putative amino acid sequences and phylogenetic tree analyses of clock genes of S. litura were highly similar to their homologs obtained from other moths examined. The data suggested that the clock genes may evolve species-specific categories based on conserved functions, such as between moths or between moths and butterflies.
Many studies on different insects have shown that circadian clock genes are involved in the regulation of some important physiological activities at various developmental stages (Groot, Reference Groot2014; Tomioka and Matsumoto, Reference Tomioka and Matsumoto2015). For example, Per mutations could affect the basic oscillator, such as the pupal eclosion rhythm and adult activity rhythm (Konopka and Benzer, Reference Konopka and Benzer1971); Vri could affect wing morphology by interacting with the biological skin growth factor decapentaplegic (dpp) signal transduction pathway (George and Terracol, Reference George and Terracol1997); Cry could regulate the feeding and metabolism of Drosophila (Seay and Thummel, Reference Seay and Thummel2011); eggshells could also be related to the regulation of insect biological clocks (Cuesta et al., Reference Cuesta, Lahiri, Lopez-Olmeda, Loosli, Foulkes and Vallone2014); Per, Tim, and Cry2 could be related to the diapause of Culex pipiens (Meuti et al., Reference Meuti, Stone, Ikeno and Denlinger2015); and the Tim gene knockout could disrupt eclosion behavior in Drosophila and affect metamorphosis in T. castaneum (Jiang et al., Reference Jiang, Yuan, Bai, Wang, Zhou and Zhu2018; Li et al., Reference Li, Yun, Yu and Li2018a). Our results showed that all clock genes could be detected at each developmental stage of S. litura, and some circadian clock genes were highly expressed in specific developmental stages, different sexes, and tissues of adults, indicating that circadian clock genes of S. litura may have similar functions to those reported above; that is, they may be involved in regulating the physiological activities of S. litura at different developmental stages from egg to adult, such as SlitCry1, four clock genes (SlitPer, SlitCry2, SlitTim1, and SlitCyc), and three genes (SlitCry2, SlitTim2, and SlitCyc) may regulate the development of eggs, adults, and some sex-related behaviors, respectively.
It is well known that the precise and complex SP communication system plays an important role in sexual communication and species differentiation in insects, especially moths (Vogt, Reference Vogt, Gilbert, Iatro and Gill2005). This system ensures that moths can perform courtship and mating rhythmically (Groot, Reference Groot2014). Previous studies have shown that female moths can use specific SP biosynthesis-related genes to precisely biosynthesize SPs at different ratios, such as the desaturase gene (Des) and fatty acyl reductase gene (FAR) (Roelofs, Reference Roelofs1995; Roelofs and Jurenka, Reference Roelofs and Jurenka1996; Merlin et al., Reference Merlin, Lucas, Rochat, François, Maïbèche-Coisne and Jacquin-Joly2007). Male moths usually use specific SP perception-related genes to accurately recognize the conspecific female SPs, such as PBPs and pheromone receptors (PRs) (Vogt, Reference Vogt, Gilbert, Iatro and Gill2005; Leal Reference Leal2013). However, whether the expression of these SP communication-related genes is rhythmic and whether they are regulated by the circadian clock is still unclear. According to previous studies (Liu et al., Reference Liu, Liu and Dong2013; Zhang et al., Reference Zhang, Yan, Liu, Jacquin-Joly, Dong and Wang2015a; Xia et al., Reference Xia, Zhang, Ding, Wang and Lofstedt2019; Zhang et al., Reference Zhang, Zhang, Zhu, Zheng, Yan, Zhu, Xu, Zhang, He, Sun, Palli, Zhang and Dong2019), nine genes (two Dess, four PRs, and three PBPs) related to SP communication in S. litura have been functionally confirmed; thus, we chose these genes and all the clock genes of S. litura for dynamic expression analysis in this study.
Previous studies confirmed that the peak of courtship and mating of S. litura adults is at 2- to 4-day-old of age (Sun et al., Reference Sun, Hu and Du2002), and our results showed that two SP biosynthesis-related genes (SlitDes5 and SlitDes11) and two clock genes (SlitPer and SlitCry2) have similar dynamic expression trends, with their peak expression levels at 2- to 4-day-old of age. Furthermore, the daily dynamic analysis of these four genes showed similar expression trends in the scotophase, and all had a peak expression before the 8th hour of the scotophase, which is just before the peak of female courtship (Sun et al., Reference Sun, Hu and Du2002). The courtship and mating of moths is a behavioral response mediated by SP signals (Ando and Yamakawa, Reference Ando and Yamakawa2011), which supports the notion that SlitDes5 and SlitDes11 are the key genes in the biosynthesis process of S. litura SPs (Zhang et al., Reference Zhang, Zhu, Fang, He, Wang, Chen, Sun, Ye, Deng and Li2015b; Xia et al., Reference Xia, Zhang, Ding, Wang and Lofstedt2019; Zhang et al., Reference Zhang, Zhang, Zhu, Zheng, Yan, Zhu, Xu, Zhang, He, Sun, Palli, Zhang and Dong2019). Therefore, we infer that SlitDes5 and SlitDes11 can promote the content of SlitDes5 and SlitDes11 by increasing the mRNA expression level before the female moths enter the courtship peak to ensure the biosynthesis of SPs. Given that the expression of Per and Cry2 is usually regulated by CLK and CYC in insects (Tomioka and Matsumoto, Reference Tomioka and Matsumoto2015), we presume that CLK and CYC may be the transcription factors for the two SlitDess, and SlitPer and SlitCry2 may indirectly regulate the expression of SlitDes5 and SlitDes11.
Perret et al. (Reference Perret, Aujard, Seguy and Schilling2003) confirmed that the olfactory bulb shows significant circadian changes in nocturnal animals. In insects, studies have shown that olfactory behavior and the expression of some olfactory genes have circadian rhythm characteristics. For example, Merlin et al. (Reference Merlin, Lucas, Rochat, François, Maïbèche-Coisne and Jacquin-Joly2007) found that the transcripts of some clock and olfactory genes cycled circadianly in the antennae of S. littoralis (Merlin et al., Reference Merlin, Lucas, Rochat, François, Maïbèche-Coisne and Jacquin-Joly2007). These also had a circadian rhythm in the expression profiles of some olfactory genes in the antennae of S. exigua, similar to their behavioral rhythm (Wan et al., Reference Wan, Qian and Du2015); Nagari et al. (Reference Nagari, Szyszka, Galizia and Bloch2017) provided evidence for circadian regulation of antennal olfactory responsiveness and odorant pulse-tracking capacity in bees or other hymenopteran insects (Nagari et al., Reference Nagari, Szyszka, Galizia and Bloch2017). According to our results, there is no similarity in the dynamic expression between SP perception-related and circadian clock genes in the MA of S. litura; however, three SP perception-related genes (SlitOR6, SlitOR13, and SlitPBP1) and four clock genes (SlitPer, SlitTim2, SlitClk, and SlitCry2) had similar daily expressions, suggesting the regulation of clock genes based on SP perception of S. litura may only occur at the male age and time of the SP perception peak. Clock genes may also participate in other physiological behaviors involving non-SP perception in MA during other periods. We also noticed that these three genes (SlitOR6, SlitOR13, and SlitPBP1) in S. litura were MA-specific or highly expressed and played a leading role in the process of SP perception (Liu et al., Reference Liu, Liu and Dong2013; Zhang et al., Reference Zhang, Yan, Liu, Jacquin-Joly, Dong and Wang2015a). These findings indicate that the expression characteristics of these genes were consistent with the rhythm of courtship and mating behaviors of S. litura and may also be regulated by the clock genes (SlitPer, SlitTim2, SlitClk, and SlitCry2). Furthermore, we found that the expression profiles of some genes (SlitCry1, SlitCry2, and SlitClk in the female PG, and SlitOR6, SlitOR13, SlitPer, SlitTim2, SlitClk, and SlitCyc in the MA) were quite different between the two photophases. Generally, moths will increase 1 day (age) after a complete scotophase, whereas adult S. litura live for only 5 or 6 days, and the changes occurring at each age may affect physiological activities in vivo. Therefore, we infer that the reason for this phenomenon may be that the increase in age leads to a change in the physiological response of S. litura in vivo. Of course, this may also be caused by other factors. We plan to use different approaches to focus on the molecular mechanisms of this difference in the future.
In conclusion, we identified and analyzed the characteristics of the sequence, genome structure, and phylogeny of all candidate circadian clock genes from S. litura based on genomic data analysis. We comprehensively compared the developmental spectrum, tissue, and temporal expression as a first step toward understanding their potential role in regulating SP communication behavior. We demonstrated that some clock genes might regulate the biosynthesis and perception of SPs by regulating the rhythmic expression of SP communication-related genes. These findings will help us elucidate the molecular mechanism of circadian clock genes involved in the regulation of sexual communication behavior and provide potential target genes for developing effective sexual communication disruptors in the future.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485321000559
Financial support
This work was supported by the National Natural Science Foundation of China (31970456), Anhui Provincial Natural Science Foundation (2008085MC63), and Natural Science Fund of Education Department of Anhui Province, China (KJ2019A0586 and KJ2019B09), Anhui College Students' Innovation and Entrepreneurship Training Program (202010373096 and S202013620012).
Conflict of interest
None.
Ethical standards
This article does not include any study on human participants or animals performed by any of the authors.











