1. Introduction
Medical education requires trainees and practising doctors to develop expertise in diagnosis or clinical reasoning. These skills are traditionally acquired through clinical practise, and they may be enhanced with the help of simulations with mannequins, role-playing games or virtual patients (VPs) (Rombauts Reference Rombauts2014). More broadly, the literature uses the term virtual patient to refer to simulations such as case presentations, interactive patient scenarios, high-fidelity mannequins, VP games, high-fidelity software simulations, human standardised patients – who are actors playing the role of interviewed patients paid for educational purposes – or virtual standardised patients (Talbot et al. Reference Talbot, Sagae, John and Rizzo2012a). VPs allow health professionals to practise their skills by interacting with a software ‘that simulates real-life scenarios’ (Cook et al. Reference Cook, Erwin and Triola2010). In our work, VP refers to virtual standardised patients. For the last few decades, VPs have allowed doctors to train clinical and history-taking skills through simulated scenarios in digital environments (Ellaway et al. Reference Ellaway, Candler, Greene and Smothers2006; Danforth et al. Reference Danforth, Procter, Chen, Johnson and Heller2009).
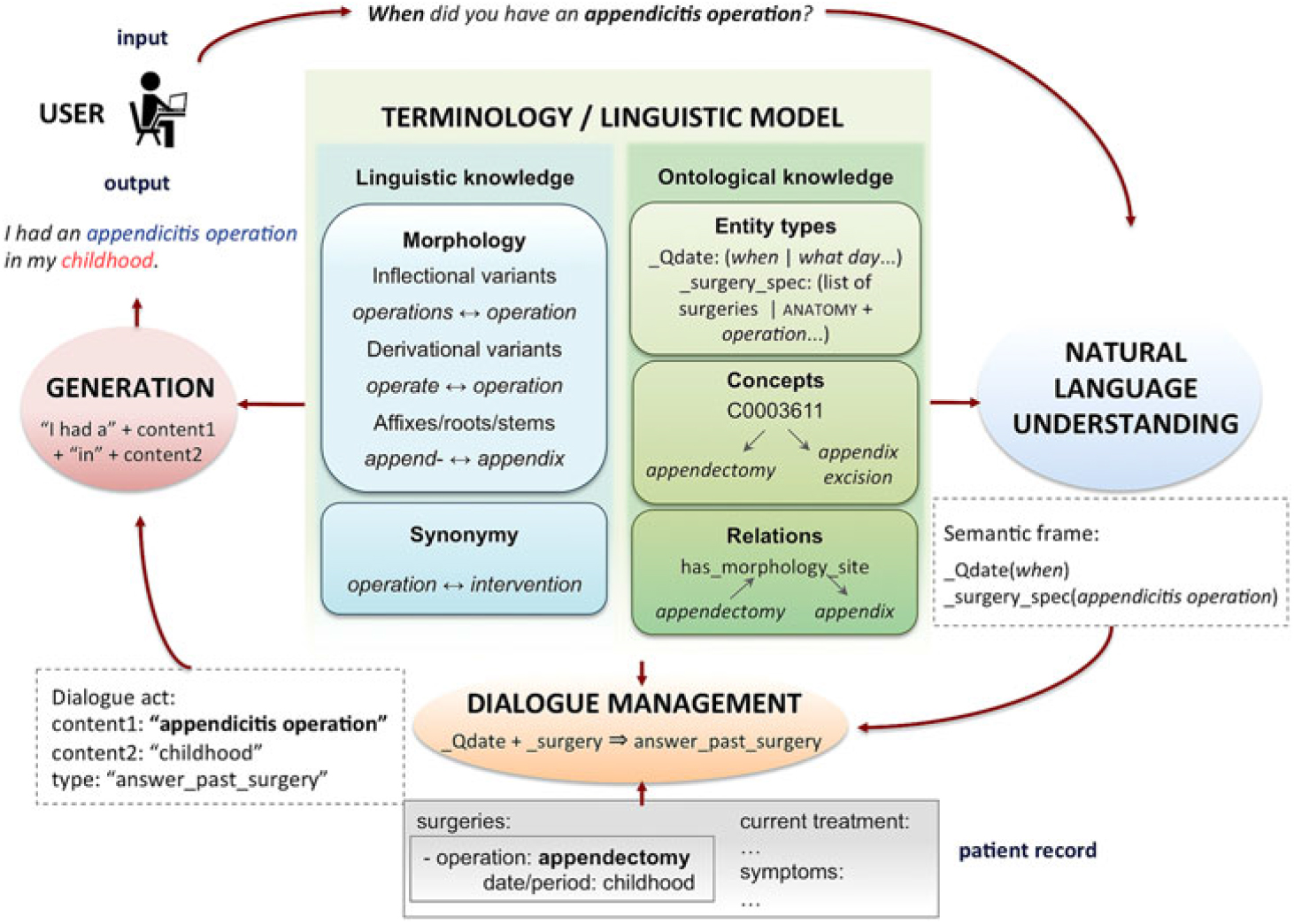
Interactivity with a VP might be enhanced through a dialogue system, but such a component needs to address several phenomena to achieve a natural, user-friendly dialogue (Figure 1). As shown, medical doctors tend to begin by eliciting initial clues from the patient by using broad questions. Then, they use follow-up questions to focus on specific details. The system needs to deal with this behaviour by processing context information (ellipsis and anaphora) and updating its information state, so that it avoids providing redundant answers. In addition, term variants referring to the same concept need to be mapped accurately (e.g. hypertension ↔ high blood pressure) by means of linguistic and terminological knowledge.

Figure 1. Sample dialogue (D: doctor; P: patient, simulated by the system) and relation to the patient record. Phenomena to be addressed are shown in bold: discourse phenomena, linguistic variation (blue circles) and termino-ontological variation (green boxes). The patient record structure is simplified due to space constraints. We show real replies of the current version of the French system, but we translated them to English.
This work describes our endeavour to create a dialogue system featuring unconstrained natural language interaction in a simulated consultation with a VP. We built this system to simulate history-taking in an educational software featuring an animated avatar with text-to-speech (Figure 2) and allowing students to simulate a physical exam. This project was developed in collaboration with a medical team specialised in simulation-based medical education at Angers University Hospital (CHU d’ Angers) and several companies (Interaction Healthcare/SimforHealth, Voxygen and VIDAL®). Within this partnership,Footnote a each group deployed their know-how: (1) to provide the system with advanced dialogue capabilities when users interview each VP: this was our contribution and is the background for the present paper; (2) to endow each VP with text-to-speech and minor gestures; (3) to develop a dedicated interface for instructors to create unlimited avatars and patient records; (4) to manage the evaluation with potential end-users who tested the system; and (5) to provide domain texts and data for the task. In this project, which spanned over three years, four computational linguists and one software engineer (about 4.5 person-years) were involved to create the dialogue system.

Figure 2. Sample of dialogue with a VP avatar.
In this context, the medical team set specific design constraints:
• The system should manage a dialogue for unseen cases without being extended manually. Achieving high vocabulary coverage allows trainees to interact with VPs in a wide range of clinical conditions. The items to which a doctor must pay attention vary widely across cases and specialities. Practising in front of a variety of VPs should therefore be a good exercise to train students in medical history-taking. Certainly, practising the history-taking skills with many cases needs to be accompanied by quality scenarios carefully designed by medical instructors to provide feedback and meaningful learning opportunities.
• The VP should provide answers based on the actual content of the patient record, that is, the information representing the patient’s health or findings (e.g. disease history or medication, Section 3.1). This brings up the need for providing correct replies, an aspect we refer to as correctness. By this, we mean that the simulated patient should not invent information that is not present in the patient record (faithfulness). Related to that aspect, the system should not omit data from the record (exhaustiveness of information). In our project, it was deemed that the system should provide all data available with regard to a given question. This design criterion seemed more adequate to avoid adding more difficulties to other possible sources of miscommunication arising with an artificial agent. Note that this was a pedagogical choice for a first version of the system: a VP could indeed simulate situations in real life where the patient lies or makes mistakes in recalling the health record. Medical students should keep that in mind when conducting a medical history.
We can add the following observations related to the task and domain:
• The conversation does not take place in an open context but in a specific domain. The medical history-taking is a focused task, defined by the number and nature of the topics that are to be processed (Section 3.2).
• The medical domain involves a large concept space with a multitude of term variants. This requires entity linking (also called entity normalisation) in order to query and match input terms with those in the patient record. Our goal is to develop a system that, first, replies correctly (correctness), and second, can work with an unlimited number of cases and medical specialities (high vocabulary coverage). A method to evaluate these aspects at the natural language understanding (NLU) step is to compute precision and recall: precision is related to correctness, whereas recall depends on vocabulary coverage. When evaluating dialogue management, correctness is a key aspect.
• A lexical simplification process is needed to simulate natural replies, that is, using terms according to a patient viewpoint (e.g. saying appendix operation instead of the professional term appendectomy written in the patient record).
• Pre-existing dialogue data for the task and domain are not available, hence the system cannot be designed with a data-intensive approach. In this sense, another contribution of our work is making available for the community a corpus of dialogue data collected during this project.Footnote b
Herein we explain how we address these requirements in our dialogue system and the challenges involved. To handle the needs of the domain and the diversity of its concepts, we applied a comprehensive approach to terminology collection. To the best of our knowledge, we integrated a larger volume of resources than in standard task-oriented conversational agents (see Section 3). We propose a framework for managing the linguistic and terminological needs in such a task, with different levels of knowledge representation, keeping coherence across components. The difficulty of term detection in our task motivates our approach based on rich terminological resources, which complement a frame- and rule-based dialogue management. The proposed methods aim to enhance the system’s capability to adapt to new cases in a wide range of medical specialities and detect rare and unseen vocabulary items for a successful interaction. To show the extent to which it succeeds in doing so in a real-use scenario, we conducted evaluations which include user interactions (n = 71) with 35 different VP cases from 18 different medical specialities, and an assessment of vocabulary coverage on 169 new cases.
We developed French, English and Spanish versions of the system. We report here work related to the French version because it is the only one evaluated to date. In the remainder of the paper, we first review the approaches to dialogue systems and to interactions with simulated VPs (Section 2). We present our termino-ontological and linguistic models in Section 3 and summarise the architecture of our system in Section 4. We present the evaluation of resources and discuss its results in Section 5, then conclude in Section 6.
2. Related work
2.1 Approaches to dialogue systems
A textual or spoken dialogue system involves several modules, which typically include NLU, speech recognition, generation, speech synthesis and a dialogue manager. More components may be added to correct input errors, query a database or search in a document collection, if the task requires it.
Different approaches exist for dialogue management (Jokinen and McTear Reference Jokinen and McTear2009): finite-state or graph-based approaches (Cole Reference Cole1999); frame-based techniques (McTear et al. Reference McTear, O’Neill, Hanna and Liu2005; van Schooten et al. Reference van Schooten, Rosset, Galibert, Max, op den Akker and Illouz2007); statistics-based approaches such as reinforcement learning (Sutton and Barto Reference Sutton and Barto1998) or partially observable Markov decision processes (POMDP) (Young Reference Young2006); and neural-network-based approaches, which have recently been reviewed (Celikyilmaz, Deng and Hakkani-Tur Reference Celikyilmaz, Deng, Hakkani-Tur, Deng and Liu2017).
Due to the lack of existing dialogue corpora for this domain, statistical or neural approaches are not applicable to the design of our VP dialogue system. Our approach aims at endowing the system with capabilities to manage a comprehensive range of aspects of a dialogue task. This involves processing input so that it is understood in the context of previous questions and answers: interpreting discourse phenomena (such as co-reference or ellipsis) and maintaining the global dialogue state. This contrasts with question-answering approaches, designed to answer independent questions, in which a system analyses the user’s natural language questions and produces a natural language answer, but commonly without processing of dialogue history (Talbot et al. Reference Talbot, Kalisch, Christoffersen, Lucas and Forbell2016; Maicher et al. Reference Maicher, Danforth, Price, Zimmerman, Wilcox, Liston, Cronau, Belknap, Ledford, Way, Post, Macerollo and Rizer2017; Jin et al. Reference Jin, White, Jaffe, Zimmerman and Danforth2017).
2.2 User interaction in health care applications
Interactive systems for health care applications address patient education and counselling (Giorgino et al. Reference Giorgino, Azzini, Rognoni, Quaglini, Stefanelli, Gretter and Falavigna2005; Bickmore Reference Bickmore2015) or support to practitioners (Beveridge and Fox Reference Beveridge and Fox2006); a review is reported in Bickmore and Giorgino (Reference Bickmore and Giorgino2006). We focus on a dialogue with a VP in an educational context. Development challenges are similar across health dialogue systems (Hoxha and Weng Reference Hoxha and Weng2016). Literature reviews on VPs are available (Cook et al. Reference Cook, Erwin and Triola2010; Kenny and Parsons Reference Kenny, Parsons, Perez-Marín and Pascual Nieto2011; Salazar et al. Reference Salazar, Eisman Cabeza, Castro Peña and Zurita2012; Rossen and Lok Reference Rossen and Lok2012; Lelardeux et al. Reference Lelardeux, Panzoli, Alvarez, Galaup and Lagarrigue2013; Rombauts Reference Rombauts2014).
A key strategy for enhancing the simulation is to provide realistic user interaction. Integrating natural language interaction into a VP system requires managing domain terms – for example, by formalising ontological concepts (Nirenburg et al. Reference Nirenburg, Beale, McShane, Jarrell and Fantry2008a) – and NLU. The NLU unit may rely on text meaning representations for resolving paraphrases (Nirenburg et al. Reference Nirenburg, McShane and Beale2009), a corpus of questions and replies curated by experts (Kenny et al. Reference Kenny, Parsons, Gratch, Rizzo, Prendinger, Lester and Ishizuka2008) or canned questions and answers (Benedict Reference Benedict2010; Siregard, Julen and Lessard Reference Siregard, Julen and Lessard2013). The i-Human Patients® systemFootnote c allows users to choose the questions to ask to the VP, whose answers are parametrised by a patient record.
Few systems allow natural language input. As far as we can tell, current tools with natural language interaction are available for practising patient assessment and diagnosis (Hubal et al. Reference Hubal, Kizakevich, Guinn, Merino and West2000) – for example, in a paediatric scenario (Hubal et al. Reference Hubal, Deterding, Frank, Schwetzke and Kizakevich2003) – and clinical history-taking and communication skills – for example, in a case of acute abdominal pain (Stevens et al. Reference Stevens, Hernandez, Johnsen, Dickerson, Raij, Harrison, DiPietro, Allen, Ferdig and Foti2006); in a psychiatric consultation (Kenny et al. Reference Kenny, Parsons, Gratch, Rizzo, Prendinger, Lester and Ishizuka2008); or in a case of back pain (Gokcen et al. Reference Gokcen, Jaffe, Erdmann, White, Danforth and Calzolari2016; Maicher et al. Reference Maicher, Danforth, Price, Zimmerman, Wilcox, Liston, Cronau, Belknap, Ledford, Way, Post, Macerollo and Rizer2017).Footnote d Kenny and his team reported using 459 question variants mapped to 116 responses related to a post-traumatic stress disorder case (Kenny et al. Reference Kenny, Parsons, Gratch, Rizzo, Prendinger, Lester and Ishizuka2008). Gokcen and colleagues’ system partially relies on manually annotated data – to date, 104 dialogues and 5347 total turns (Gokcen et al. Reference Gokcen, Jaffe, Erdmann, White, Danforth and Calzolari2016). The Maryland Virtual Patient, which simulates seven types of oesophageal diseases, uses a lexicon covering over 30,000 word senses and an ontology of more than 9000 concepts (Nirenburg et al. Reference Nirenburg, McShane, Beale and Jarrell2008b).
These interactive systems seem to be case specific; that is, they treat a limited number of cases. As far as we know, Talbot et al. (Reference Talbot, Kalisch, Christoffersen, Lucas and Forbell2016) developed one of the few natural language interaction systems trained to cope with different clinical cases in the English language (e.g. ear pain, psychiatry and gastroenterology).Footnote e It relies on a medical taxonomy of 700 questions and statements and a supervised machine-learning model trained on over 10,000 training examples. For their part, the Virtual Patients Group (VPG, a consortium of North-American universities) also envisages a robust natural language interaction system. The VPG’s platform Virtual Patient FactoryFootnote f allows users to create new cases and interact with virtual humans. Application scenarios range from psychiatry to pharmacy. To develop the NLU component, they used the human-centred distributed conversational modelling (HDCM) technique (Rossen, Lind and Lok Reference Rossen, Lind, Lok and Ruttkay2009), a crowd-sourcing methodology for collecting the corpus used to feed the system. Their method relies on a tight collaboration between VP developers and medical experts, a workflow that we specifically aim to bypass to make the system much more easily extensible to new cases.
Lastly, neural approaches are being explored for the NLU component in VP systems (Datta et al. Reference Datta, Brashers, Owen, White and Barnes2016; Jin et al. Reference Jin, White, Jaffe, Zimmerman and Danforth2017). These are data-intensive methods and can be set up once enough data are collected from real interactions.
We applied a knowledge-based approach, mostly rule and frame based, because of the lack of available dialogue and domain data to train a machine-learning system. Due to the magnitude of the terminology in the medical domain, we also rely on rich terminological resources, which led us to give special care to the design of language resources management.
3. Models
Given a clinical case, the medical trainee will ask questions about various facets of the patient record, referring to entity types and concepts through domain terms. In this section, we present the models designed to create our VP dialogue system. First, the patient record model (Section 3.1) defines the patient’s health status. Second, the knowledge model of the dialogue task (Section 3.2) defines the range of questions types, entity types and dialogue acts. Third, the linguistic and termino-ontological model (Section 3.3) defines domain relations and concepts and manages linguistic variation.
3.1 Patient record model
We first need to specify what type of information is available to the dialogue system about the patient’s state. This is the aim of the patient record model, from which the dialogue system will provide information about the specific patient it embodies. This model is similar to those that underlie electronic health records and was refined iteratively when the first cases were created.
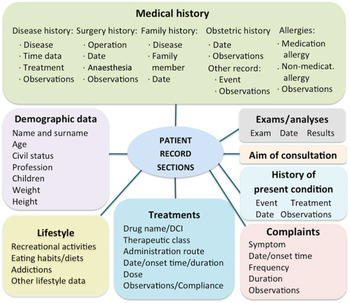
The VP is specified in a clinical record typically authored by a medical trainer, who aims at teaching students how to handle a given case. The clinical record describes common data found in patient records, structured into sections (e.g. Medical history or Current treatment) and lower-level subsections (e.g. Medical history has a subsection on Family history; Figure 3). Some basic elements can have associated attributes (e.g. the dose and frequency of a medical treatment). Most elements and attributes consist of free text. An example of a clinical record is shown in Table 1.
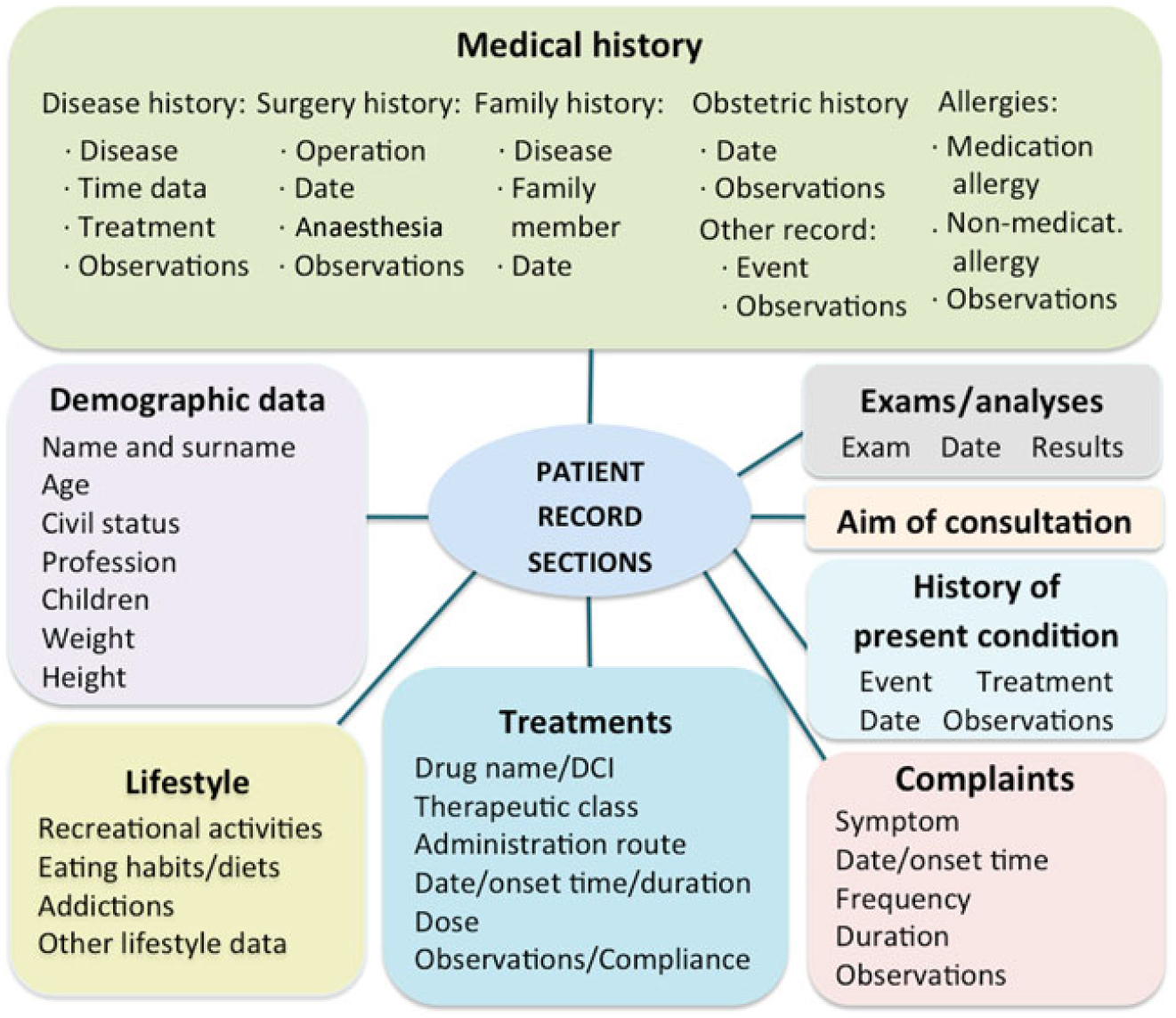
Figure 3. Patient record model.
Table 1. Sample clinical record: the format used is YAML

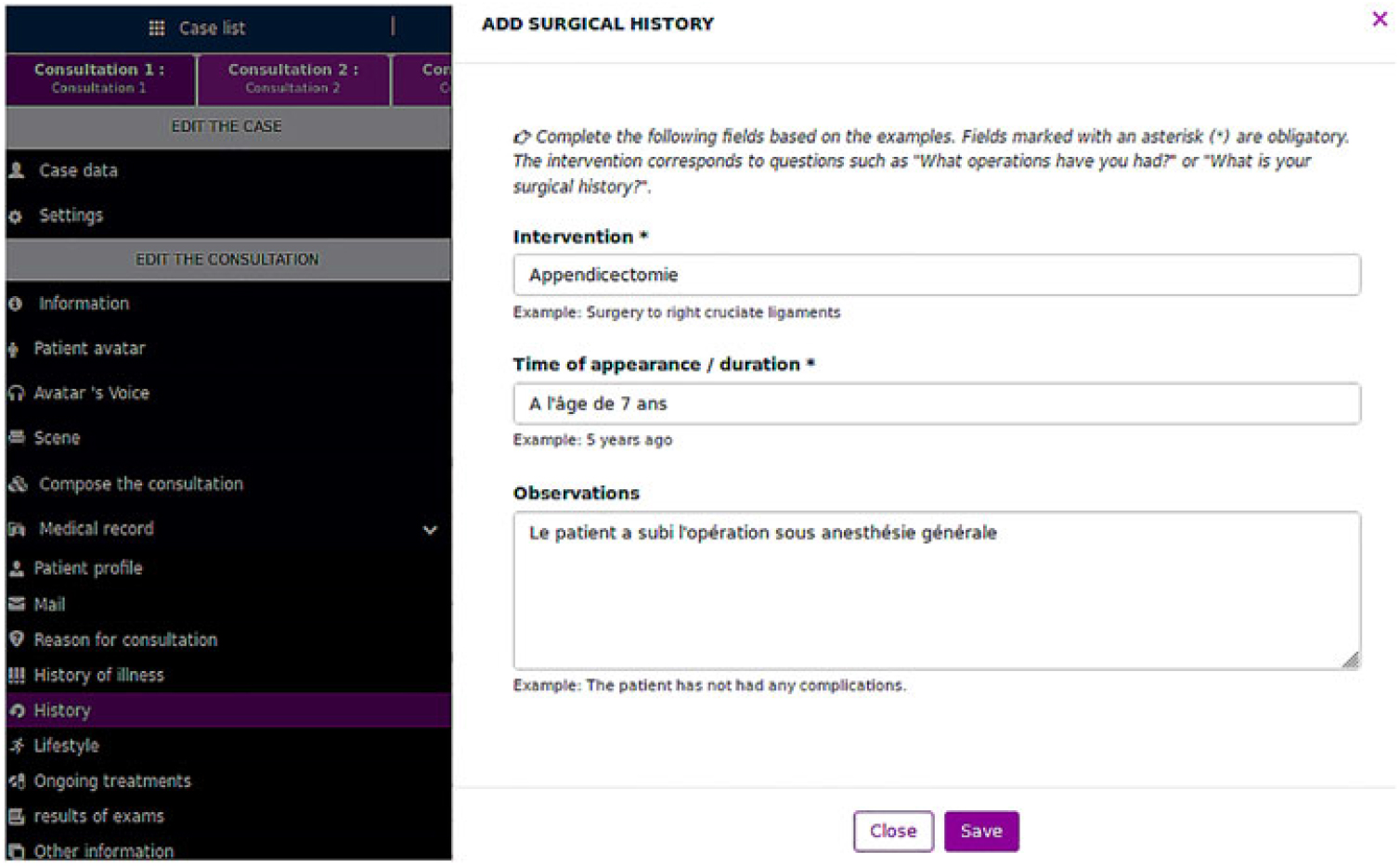
Medical instructors prepared patient records through a dedicated interface (Figure 4). A preprocessing module extracts information from some text fields, for example, time data from the symptoms (complaints) field. The system most often reuses text from the patient record to generate replies, which may make them less natural.

Figure 4. Dedicated interface to input data on the VP record.
3.2 Knowledge model for the dialogue task
Our system operates in the context of the anamnesis stage of a consultation scenario – that is, the medical history-taking step to collect diagnostic information from a patient. Therefore, the system must be capable of analysing and replying to common questions (related to symptoms or treatments). The system also needs to process broader designated topics that are necessary to conduct history-taking, namely, demographic data (patient’s name and surname, civil status, age, profession or family status) and questions on patient’s lifestyle (recreational activities, eating habits, social life or family life).
The knowledge model for the dialogue task defines:
• the range of topics that might be addressed during the clinical anamnesis, and the relation to each record section (as they were grouped in our project);
• the question types to be processed in the dialogue manager, as well as the sections in the VP record associated to these question types;
• the entity types to annotate in the user input by the NLU module (Section 3.3.2);
• dialogue acts, which define the function of the user input (e.g. greeting, acknowledging) analysed by the NLU module, and acts that define the reply type and content to be output by the natural language generation (NLG) module (e.g. inform_symptom_duration).
A sample of the model with regard to symptoms is described in Figure 5.

Figure 5. Knowledge model for the dialogue task (sample). The upper part shows the relation between questions on the patient record, questions types and topics, and entity types. The lower part shows how system components instantiate this model. NLU: natural language understanding; NLG: natural language generation.
We designed the model based upon the following sources. We collected questions used in a patient–doctor consultation scenario. We used 30 audio recordings with human standardised patients, who simulated consultations on anaesthesiology, hypertension and pneumopathy. Several actors were recorded for each case, which allowed us to obtain varied versions of history-taking for the same case.Footnote g We transcribed and analysed the recordings to detect interaction patterns. We also gathered questions from guides for clinical examination used by practitioners (Bates and Bickley Reference Bates and Bickley2014; Epstein et al. Reference Epstein, Perkin, Cookson and de Bono2015) and from resources for medical translation (Coudé, Coudé and Kassmann Reference Coudé, Coudé and Kassmann2011; Pastore Reference Pastore2015).Footnote h
For entity types and dialogue acts, we defined 149 different labels: 62 entity types, 70 question types and 17 dialogue acts (e.g. greetings). A total of 52 labels are used for medical entity types and 10 for non-specifically medical entity types (e.g. frequency). A total of 56 labels are used for medical question types (e.g. Qsymptom) and 14 for general question types (e.g. for dates, Qdate, when; or cause, Qwhy, why). Medical entity and question types are related to patient record sections: lifestyle (16.7% of labels), medical history/symptoms (60.1%), treatments (8.3%), clinical examinations/analyses (4.6%) and demographic data (10.2%).
The knowledge model for the dialogue task was defined and refined in an iterative process during development: evaluators periodically interacted with the system, and a computational linguist analysed the logs to improve the questions types, the labels of entity types and the missing terms used in the interaction. The process extended over 7 iterations and about 24 months. We report the domain sources used for the lists of entity types in Table 3 (Section 3.3.2).
3.3 Linguistic and termino-ontological model
Medical terminology brings up multiple processing difficulties. To illustrate their order of magnitude, let us first introduce the Unified Medical Language System® (hereafter, UMLS®) (Bodenreider Reference Bodenreider2004). The UMLS MetaThesaurus® is a large multilingual source of medical terminologies and ontological knowledge that come from close to 200 thesauri. In a similar way to how WordNet encodes synsets, the UMLS MetaThesaurus encodes concepts from different vocabulary sources with Concept Unique Identifiers (CUIs). CUIs map concepts and terms across multiple terminologies. Likewise, the UMLS Semantic Network® compiles semantic relations (e.g. is_a or caused_by) from the source ontologies. Terms in the UMLS are classified into 134 semantic types (STYs); for example, diabetes is a Disease or Syndrome, and fever is a Sign or Symptom. UMLS STYs are clustered into 15 semantic groups. For example, the group DISO contains various types of health conditions such as diseases, injuries, symptoms or findings. The UMLS MetaThesaurus contains 349,760 distinct French terms in the 2017AA version (4011 of the type Sign or Symptom). Terms are often nested (e.g. heart failure) and variation includes derivation (heart vs. cardiac), compounding (cardiovascular), abbreviation (MI, for myocardial infarction) and lay terms (heart attack).
The above description shows that the number of different concepts and terms in this domain is larger than in usual dialogue systems. In such a context, we needed to provide the system with resources for concepts and words so that it knows about the domain and can handle term variation to interact adequately in it.
We developed for this purpose a linguistic and termino-ontological model, which hosts structured thesauri with linguistic, terminological and ontological knowledge. It is schematised in the central block of Figure 6 (left and right panels, respectively).

Figure 6. Linguistic and termino-ontological model at the core of system architecture.
With regard to the different language versions of the system, processing term variants is more challenging for French or Spanish, due to the higher number of verb forms or gender variants compared to English. For example, we needed resources for gender and number agreement to generate grammatically correct replies; accordingly, these resources are larger for the versions in French and Spanish.
3.3.1 Overall description
The variability of natural language expressions calls for linguistic knowledge. This includes word-level information: morphological information such as inflection (e.g. kidney ↔ kidneys), derivational variants (e.g. surgery ↔ surgical), affixes and root elements (e.g. disease ↔ -pathy) and synonyms (e.g. operation ↔ surgery).
Termino-ontological knowledge defines the relations and concepts that are useful for the system to interact in the domain. The structure of thesauri is similar to that in the UMLS Metathesaurus and is organised around entity types, terms linked to concepts and relations between these concepts.
We distinguish entity types (semantic classes of the domain defined for the task, e.g. label treatment) and concepts (conceptual items related to entity types). Terms refer to concepts, and concepts are classified with entity type labels (Figure 7). For each concept, termino-ontological knowledge provides one or more terms, which are used to handle term variation (e.g. hypertension ↔ high blood pressure).

Figure 7. Relation between entity types, concepts and terms. Linguistic knowledge processes affixes or morphological variants; ontological knowledge classifies terms into entity types; UMLS terms are indexed by CUIs.
The linguistic knowledge is instantiated through language resources such as dictionaries; it has no direct link to concepts or entity types. The termino-ontological knowledge is typically instantiated by domain terminologies. Accordingly, we use different linguistic and terminological resources. There is a separate lexicon file for each component (e.g. a file for synonym variants and another for derivational variants in the linguistic model). Table 2 shows the types of variation phenomena and the resources needed for the generation, entity linking and normalisation steps. For instance, the generation step (first pane of the table) uses information stored in both the linguistic and termino-ontological models (this is detailed in Section 4.5). Here, linguistic knowledge consists of morphological information: gender, number and part of speech, as well as correspondences between specific verb forms (e.g. has ↔ have). Termino-ontological knowledge maps scientific and lay term variants (e.g. per os ↔ oral). Likewise, the entity linking and normalisation step (second pane of Table 2) uses linguistic knowledge to manage linguistic variation (see Section 3.3.3) and termino-ontological knowledge to manage terminological variation (see Section 3.3.4).
Table 2. Types of resources used in each processing step for managing linguistic and terminological variation (and examples of each type), listed according to linguistic aspects (morphology and synonymy) and termino-ontological aspects (concepts and ontology relations)

To build these lexicons, we extracted semi-automatically terms and ontology relations from the UMLS where possible. Due to the large size of the UMLS, we used the subset of its terminologies and STYs that were relevant for our task; for example, we did not need entity types such as Regulation or Law (STY T089). We also used the National Agency for the Safety of Medicines listFootnote i and extended the lexicons as needed for our task. We built two types of lexicons: lexicons with terms recorded in the UMLS (with a CUI) and other lexicons that we created with terms not recorded in the UMLS. We explained the methods to collect them in a previous work (Campillos-Llanos et al. Reference Campillos-Llanos, Bouamor, Zweigenbaum, Rosset and Calzolari2016), and we briefly describe them below.
3.3.2 Entity types
Vocabulary lists to label domain and miscellaneous entities (both mono- and multi-word items) amount to 161,878 items. Table 3 shows the correspondence between patient record sections, entity types, NLU labels, the source of the data and the sizes of the associated term lists. The counts presented do not aggregate typographical variants; for example, anti-hypertensive and antihypertensive are counted separately. We provide the source used for extracting some domain terms from the UMLS (column Source): codes are UMLS STYs (e.g. T184 for Sign or Symptom) or UMLS semantic groups. Our list of diseases has items of all DISO STYs except Sign or Symptom (T184). We specify in italics the Medical Subject Headings (MeSH) code where this terminology was used, for example, D27.505 (Pharmacological action) for drug therapeutic classes. When there was not an UMLS STY for the entity type needed, we extracted terms by using regular expressions. For example, to collect the list of allergies, we applied the expression (allerg|intolér|réaction) on UMLS terms of type Disease or Syndrome (T047), and then manually revised results. Some lists come from the same STY, but semi-automatic and manual methods were used. For example, we extracted terms for gynaecological and obstetric events from the type Disease or Syndrome. During development, after users interacted with the system, we observed that some question types were related to obstetrics entity types (e.g. miscarriage), and others related to gynaecological events (e.g. abundant menstruation). Hence we needed to distinguish between both of them and refined the scheme of entity types.
Table 3. Correspondence between VP record sections, NLU labels and lists, and sources

3.3.3 Managing linguistic variation
We address two types of linguistic variation: morphological variation (inflection and derivation) and synonymy. The former involves dealing with inflectional variants (e.g. to sweat and sweating) (Table 2, 2.1). We manage this through a general-language inflectional dictionary – we used DELA-type electronic dictionaries for French (Courtois Reference Courtois1990). Also, derivational variants may occur (e.g. impairment and impair; Table 2, 2.2). We deal with them through deverbal nouns collected from a specialised general lexicon – for French, VerbAction (Hathout et al. Reference Hathout, Namer, Dal, Hathout, Namer and Dal2002). Derivational variants of medical terms (e.g. fever ↔ feverish, thorax ↔ thoracic) come from the UMLF lexicon (Zweigenbaum et al. Reference Zweigenbaum, Baud, Burgun, Namer, Jarrousse, Grabar, Ruch, Le Duff, Forget, Douyère and Darmoni2005). Synonym relations (e.g. walk and stroll) are processed by means of general synonym lexicons (Table 2, 2.4). For French, we reused the dictionary applied in a previous project (Rosset et al. Reference Rosset, Galibert, Adda and Bilinski2008). This open-domain dictionary contains entries whose values are synonym words, for example, opération|manipulation;action;intervention.
3.3.4 Managing terminological variation
3.3.4.1 Terms referring to the same concept
For terms recorded in the UMLS (Table 2, 2.5), we can map term variants associated with the same concept by using their CUI, for example, high blood pressure and hypertension (CUI C0020538). Term variants come from the following UMLS types and groups (McCray, Burgun and Bodenreider Reference McCray, Burgun and Bodenreider2001):
• Anatomic entities (Table 2, 2.5.1): ANAT semantic group
• Health states (i.e. diseases/symptoms, Table 2, 2.5.2): DISO semantic group
• Pharmacologic substances (Table 2, 2.5.3): T121 STY
• Therapeutic/diagnostic procedures (Table 2, 2.5.4): PROC semantic group
Terms not recorded in the UMLS need special processing (Table 2, 2.6). For example, the UMLS lacks French verbs for symptoms (e.g. to perspire), which can not be mapped to noun forms (e.g. sweating, C0038990). We created ad hoc lists to link them, gathering single- and multi-word verbs/idioms and lemmatised forms. We collected them manually and iteratively: after users interacted with the system, we analysed interaction logs and added items to lexicons. With a similar procedure, we collected a secondary list for remaining lay variants (e.g. per os ↔ oral).
When other methods fail, medical affixes and roots/stems (Table 2, 2.3) help match terms with no UMLS relation. To build these resources, we adapted the lexicon of affixes and roots of DériF, a morphosemantic linguistic-based parser for processing medical terminology in French (Namer and Zweigenbaum Reference Namer and Zweigenbaum2004). This analyser decomposes terms into morphological constituents and classifies them into domain STYs. We also translated to French some neoclassical compounds (i.e. Latin or Greek prefixes, roots or suffixes, e.g. amygd-, ‘tonsil’) from the UMLS Specialist lexicon® (McCray, Srinivasan and Browne Reference McCray, Srinivasan and Browne1994). This lexicon is an English dictionary (over 200,000 entries) gathering biomedical terms and frequent words. Each entry records syntactic, morphological and orthographic information.
3.3.4.2 Using hierarchical relationships
Some dialogue contexts involve concepts with a different degree of specialisation to that in the record. In these cases, we use UMLS relationships to map general to specific concepts (or vice versa), especially relations from SNOMED CT (Table 2, 3). The Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT) (Donnelly 2005) is a clinically oriented multilingual terminology distributed by the International Health Terminology Standards Development Organisation (IHTSDO). SNOMED CT gathers codes, hierarchical concepts, relations between them and descriptions, and it is included in the UMLS.
Namely, UMLS child_of (chd) relations are used to map a type of disorder (e.g. cardiovascular disease) to a specific disease in the record (e.g. high blood pressure). Relationships are also used when term variants fail. Terms referring to classes of disorders with the pattern disease_gen + anatomy are related to their anatomical site by SNOMED CT relation has_finding_site, for example, kidney disease and glomerulosclerosis. Other SNOMED CT relations are used: has_procedure_site, to map anatomic terms and surgical procedures with pattern surgery_gen + anatomy (e.g. appendectomy ↔ appendix); and has_procedure_morphology or has_direct_morphology, to map entities referring to surgeries with the structure disease_spec + surgery_spec (hernia ↔ inguinal herniorrhaphy).
To relate symptoms or disorders to physiological functions (e.g. dyspnea and to breathe), we extracted lists of correspondences between these types of entities from UMLS terminologies – namely, ICD10, MeSH and SNOMED – together with hierarchical relationships between concepts of these types.
Table 4 reports the size of the resources for managing terminological and lexical variation: number of variants, minimum, maximum and mean values per CUI or lexical entry (mono- and multi-word items) and number of lexical entries or CUIs in each resource (for relations between CUIs, we give the number of related pairs).
Table 4. Resources for managing linguistic and terminological variation

4. Implementation
In this section, we first describe the general architecture of the system (Section 4.1). We then detail the resources and processes used in the NLU stage (Section 4.2 and Section 4.3), the dialogue manager and patient record querying (Section 4.4) and the generation step (Section 4.5).
4.1 Architecture of the dialogue system
The architecture of the system (Campillos-Llanos et al. Reference Campillos-Llanos, Bouamor, Bilinski, Ligozat, Zweigenbaum and Rosset2015) is based on the modular schema of the RITEL interactive question-answering dialogue system for open-domain information retrieval (Rosset et al. Reference Rosset, Galibert, Illouz and Max2006). The RITEL platform is an infrastructure for developing dialogue systems. We adapted the NLU engine and the processing functions for dialogue management and generation. Our contributions are the lexicon and models for the VP dialogue task. Our system has these modules:
• The terminology and linguistic management module provides the termino-ontological and linguistic knowledge needed by the various components.
• The NLU module analyses the user input. It recognises medical entities and question types, and includes spelling correction to deal with errors in user input. The user input is a turn and consists of one or more utterances. For instance, Hello. How are you doing? is a turn and consists of two utterances.
• The dialogue manager interprets the results of the NLU. Based on the entity and question labels produced by the NLU, rules are applied to determine the semantic frame that best represents the type of user question and drives the construction of a query to the record. The required information is looked for in specific sections of the record. The dialogue manager then passes the query results and a suitable reply type to the generation module. The dialogue manager takes into account and updates a dialogue history, using a shallow implementation of the information state approach (Traum and Larsson Reference Traum, Larsson, van Kuppevelt and Smith2003). This history keeps track of the interaction at each move and is used to process ellipsis and anaphora (see examples in Figure 1). This is an important device to manage longer, more natural conversations than what a series of independent question-answer turns would provide.
• The NLG module creates the output utterances. Instead of using predefined replies, the system relies on templates to generate new answers according to the contents of the patient record.
Figure 8 illustrates how an input question is analysed and processed to output a reply according to the VP record. After the spelling correction, the NLU module detects the terms in the user input and annotates the question and entity types. Then, the dialogue manager processes the semantic frame filled with the entity types in the input. The terminology management module looks for the corresponding data in the VP record, performing entity linking or normalisation if needed. If these data are found, the lexical module looks for a lay variant of the term (e.g. appendicitis operation for appendectomy). Lastly, the generation module applies the type of reply that corresponds, and the record items are instantiated in the generation template.

Figure 8. Example of question-reply processing.
4.2 NLU: Spelling correction
For each out-of-vocabulary (OOV) word, we attempt spelling correction. To handle common types of misspellings, we implemented a correction algorithm based upon Norvig (Reference Norvig2007). We use morphological information, word length and corpus frequency of each word in the system together with edit-distance metrics to choose the most likely correction. The spelling corrector relies on a dictionary gathering all mono-word terms in the system. Each word form corresponding to the same lemma has its own entry (e.g. tension and tensions). The current version contains 659,720 word forms.
We used a domain corpus to develop the dictionary of the spelling corrector. The VIDAL® company provided for the project its dictionary of medical drugs, which is a reference book used by health professionals and is copyrighted (therefore, not freely available).Footnote j Texts include data on common disorders and medications in French, and the corpus size amounts to 7,678,363 tokens.
Spelling correction is a useful component but is not the focus of the present paper, hence we do not expand further on its description.
4.3 NLU: Entity recognition and semantic annotation
The NLU module performs the following tasks:
• Entity recognition and semantic annotation: we use rules and domain lists and gazetteers semi-automatically curated for each entity type. More details on these lists were provided in Section 3.3 and Table 3.
• Dialogue act and question-type annotation: we use rules to label conversational dialogue acts (e.g. greetings) and classify question types (e.g. yes–no questions bear label Qyesno).
In this step, we use Wmatch rules (van Schooten et al. Reference van Schooten, Rosset, Galibert, Max, op den Akker and Illouz2007; Galibert Reference Galibert2009). Wmatch is a regular expression engine of words for natural language processing. It uses domain lists to detect words in user input and allows defining local contexts for matching and semantic categorisation. Each of the 149 entity types, dialogue acts and question types is defined by a grammar that is expressed by a combination of abstract rules and gazetteers. Each grammar generates a complex graph that is used ultimately at run time. Table 5 shows sample rules. Two computational linguists, with the expertise of a senior researcher, developed the rules in an iterative process of analysing interaction logs and refining matching contexts.
Table 5. NLU rules (actual code) for dialogue acts (greetings and acknowledging), questions on hours, and entity types (diseases and surgeries). Labels start with the _ character; & indicates a sub-expression that can be reused in different locations; | indicates alternation; ˄, start of string; <!, left negative lookahead; ?!, right negative lookahead; ?:, non-substituting grouping;.{0,n}: 0-n matching; _∼, word lemma

With regard to medical entity types, we distinguish two levels of specialisation: general (the top-level entity types of the domain, such as surgery_gen) and specific (the descendants of these top-level types, such as surgery_spec). Patrick and Li’s taxonomy also differentiates general and specific clinical questions (Patrick and Li Reference Patrick and Li2012), but we did not adapt it to our knowledge model for the dialogue task due to the different application setting.
Table 6. Types of input questions

We consider two variants of question types: Wh- questions (open questions) and yes–no questions (polarity questions) (Quirk et al. Reference Quirk, Crystal, Greenbaum, Leech and Svartvik1985). This distinction is needed to determine the type of reply (i.e., yes–no questions are replied with yes or no).
The resources used in the NLU step vary according to the type of entities in input questions (see Table 6):
• Questions on general entities: Lists of top-level entities (i.e. towards the top of the hierarchy or ontology) referring to a entity type in the domain, for example, operation belongs to the entity type surgery_gen, and disorder, to the entity type disease_gen.
• Questions on specific entities: specific entities referring to a more detailed entity type in the domain. Two types of queried entities may appear depending on the type of question topic:
- Classes of entities, for example, cardiovascular disease (disease_spec).
- Subclasses of entities, for example, appendectomy (surgery_spec) or hypertension (disease_spec).
In the NLU step, lists and rules aim at balancing both precision and recall to consider different term and question structures or spelling variants. To increase precision, we needed comprehensive lists of terms and rules defining precise matching contexts. To improve recall, we expanded term lists (e.g. by including frequently misspelled words) and relaxed the context of some rules – the less specific the context, the higher the recall. During system development, we removed noisy terms in lists and fine-tuned greedy matching rules.
4.4 Dialogue manager and patient record querying
At each dialogue move, the dialogue manager interacts with the lexical modules. First, the dialogue manager processes user input according to the semantic frame from the NLU step. In addition, the information state module updates the input content representation dynamically according to the current dialogue state. The reference of an anaphoric pronoun or an elliptic element is interpreted according to the previous dialogue state. For example, in the sample dialogue of Figure 1, the system interprets the ellipsis of the medical term in since when? as the symptom expressed in the previous reply (shortness of breath). This allows the system to manage the semantic interpretation of user input in context.
To query the record, inflected forms of entities are transformed to a base or canonical form (e.g. the singular noun or the infinitive verb form) by using the lexicons in the linguistic model. Medical entity verbs (e.g. Have you bled?) undergo some steps of lemmatisation (bled → bleed) before the base form is mapped to any variant (e.g. bleed → hemorrhage). Multiword entities also need another step to remove some pronouns and obtain a canonical form, for example, Respirez-vous avec difficulté? (‘Are you breathing with difficulty?’) is reduced to the base form respirer avec difficulté (‘breathe with difficulty’); then, this form can be mapped to a mono- or multi-word variant term in the patient record (e.g. shortness of breath or dyspnoea).
In the patient record query step, postprocessed entities are dynamically looked for in the record. The dialogue manager uses the entity type to restrict the search for data in the corresponding record section. For example, a question on a disease is looked up in the section concerning disease history.
There is a continuum between types of entities and question types (as exposed in Section 4.3). Their nature (general or specific) affects the size of processes and resources for querying the record. At one extreme, questions on general entities only require an accurate identification of the entity type. For example, a question such as What diseases do you have? requires identifying diseases as a generic term (label disease_gen). At the other extreme, questions on specific entities also demand entity linking (also called entity normalisation) to check whether the input entity and that in the record refer to the same concept. For example, a question such as Do you have tension problems? requires labelling tension as symptom and managing term variants when checking these data in the record (e.g. tension problems ↔ high blood pressure). Questions on a class of entities require matching this class with any of its subclasses in the record. If an user asks a question such as Do you have cardiovascular diseases?, which contains a term referring to a broad class, we need to map it to a specific disease in the record (e.g. high blood pressure, a subclass of cardiovascular disease). To do so, we use ontological relations.
Methods for entity linking use exact or approximate match (Levenshtein Reference Levenshtein1966) or any of the resources defined in the termino-ontological model. The specific lexicons and/or ontology knowledge to be used rely on the entity type of each term. Terms whose entity types are related to pathologies (e.g. label disease_spec or symptom) are looked up in lexicons of term variants extracted from the UMLS DISO group. That way, hypertension can be mapped to high blood pressure or hypertensive disorder. Likewise, terms belonging to procedure entity types (e.g. appendectomy, label surgery_spec) are looked up in lexicons with variants extracted from the UMLS PROC group. The input terms to be matched with terms in our lexicons belong to the same entity type; variants are not expected to be found among terms of other entity types. This restriction of the search space speeds up the dictionary look-up process. By focusing on the relevant parts of the record, the correction of answers is also expected to increase. Algorithms 1 and 2 are pseudocode examples of how these queries are implemented.
In this step, the correction of answers depends on the ability of the system to map input terms to items in the patient record. This in turn depends on the coverage and quality of the linguistic and termino-ontological resources of the system.
4.5 Generation
Resources for generating replies cover three types of information:
• Linguistic data for gender/number agreement, for example, fever is feminine in French. We use DELAS-type (Courtois Reference Courtois1990) dictionaries with inflectional information (Table 2, 1.1).
• Correspondences between third and first person verb forms to output the content expressed in the record (in 3rd person) with the patient’s viewpoint (1st person), for example, The patient has a fever → I have a fever. We clustered pairs of verb forms from the mentioned dictionaries (Table 2, 1.2).
• Lay variants of terms, for example, appendectomy → appendicitis operation. These were selected by processing domain corpora of different degrees of technicality (Bouamor et al. Reference Bouamor, Campillos-Llanos, Ligozat, Rosset, Zweigenbaum and Calzolari2016) and manual revision (Table 2, 1.3 and 1.4).
Algorithm 1 Pseudocode of function to match terms through UMLS CUIs. The function returns True when an input term and a term in the patient record refer to the same UMLS concept. Dictionaries in the termino-ontological model are selected according to a semantic code (ANAT, DISO, PROC or T121) corresponding to the input entity type.
The linguistic model is used to get the lemma of the input word form.
5. Evaluation methods and results
We present our evaluation goals and criteria (Section 5.1) and explain how we gathered evaluation data (Section 5.2). Next, we detail our evaluation methods and results for different aspects, and we end with a discussion of results (Section 5.8).
5.1 Overview of evaluation principles
One of the difficulties in evaluating dialogue systems lies in the lack of benchmarks and comparable or agreed standards (Paek Reference Paek2001). Frameworks such as PARADISE (Walker et al. Reference Walker, Litman, Kamm and Abella1997) established a foundational methodology, especially with regard to distinguishing objective and subjective metrics – or performance and usability (Roy and Graham Reference Roy and Graham2008). Human judgements on dialogue performance are thus relevant and necessary to complement other measures.
We designed and ran both quantitative and qualitative evaluations of system performance with a focus on its vocabulary coverage. Evaluating at these two levels provides us with an overall picture of how objective metrics reflect subjective assessments (Paek Reference Paek2001). More specifically, we performed the following evaluations:
• A quantitative evaluation of the NLU unit (Section 5.3).
• A quantitative evaluation of dialogue management, that is, dialogue control and context inference (Section 5.4).
Algorithm 2 Pseudocode of function to query surgery terms in the patient record. The function returns True when an input term is found in the patient record. Dictionaries and ontology relations are used from the termino-ontological model. Function Match_Term_through_CUI (see algorithm 1) is used to match terms through UMLS CUIs.
The linguistic model is used to match terms through affixes and roots.
• A qualitative evaluation of the overall functioning of the system and of its usability (end-user satisfaction) (Section 5.5).
• A quantitative evaluation of the system’s vocabulary coverage with regard to processing new cases (Section 5.6).
• A qualitative evaluation of vocabulary usage in the task (Section 5.7).
5.2 Collection of interaction data
During system development, we collected interaction data by having computer science students and researchers (n = 32) interact with the system (3 VP cases) and evaluate it through an online interface and questionnaire.Footnote k For the evaluation presented here, in the following rounds of tests, medical students and doctors (n = 39) interacted freely with the system and then evaluated it. We used 35 different VP cases; each case was tested by an average of 3.74 users (±2.8; minimum number of different users per case = 1; maximum = 13). We gave instructions concerning the types of dialogue acts the system can process (e.g. avoid instructions or out-of-task requests such as Give me your telephone number). Table 7 includes a sample of the instructions provided. Note that we did not provide examples of question formulations, removing the risk of priming effects.
The data reported in this evaluation were collected between March 2016 and February 2018. During the development tests with a small set of cases, we collected from computer science students and researchers around 1987 pairs of turns, that is, user input and system reply (a total of 3756 turns, 11,960 tokens in user input). Users with this profile have more knowledge of the human–computer interaction limits and helped us improving system’s response behaviour. Dialogues they conducted provided us with variants of question types before real end-users evaluated the system. For the evaluation presented here, we gathered from 39 medical doctors a total of 8078 turns in 131 interaction dialogues (21,986 tokens of user input and 21,921 tokens in replies). After manually inspecting our data, we removed 149 pairs of turns (3.7%) corresponding to out-of-task questions (e.g. What is your favourite colour?) or declarative statements the system is not expected to answer (e.g. Please give me reasonable replies). We are aware that some types of declarative statements are important in doctor–patient interactions, for example, to show empathy or counsel the patient (e.g. You should stop smoking). The current version of the system includes rules to process some declarative statements related to the patient’s additive behaviour. Table 8 breaks down our evaluation data (all collected turns) corresponding to medical users. Note that in some cases the system did not reply due to processing errors of the dialogue manager.
Table 7. Instructions available on the evaluation interface

Table 8. Data collected during the evaluation by medical doctors

#D: count of dialogues; #T: count of turns; #T/D: average turns per dialogue; #W: count of words; #W/D: average words per dialogue; stdev: standard deviation
5.3 Quantitative evaluation of NLU
5.3.1 Methods
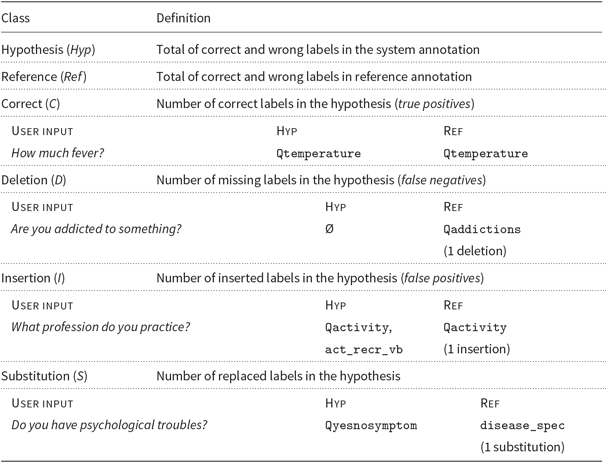
The NLU module analyses the user input and as a result provides labels for its semantic analysis. To evaluate it, we used the standard metrics of Precision, Recall, F-measure and Slot Error Rate (SER) (Makhoul et al. Reference Makhoul, Kubala, Schwartz and Weischedel1999). These were computed by counting the number of correct and incorrect entity types labelled; for wrong labels, we counted insertions (I), deletions (D) and substitutions (S) (Table 9). We also computed the Sentence Error Rate (SeER), which is the proportion of sentences with at least one error. The definitions of all these metrics are given in Table 10. A better performance is normally reflected in higher P, R and F-measures and lower SER and SeER values.
Table 9. Evaluation schema for the NLU module (with examples)
We also evaluated the spelling correction module by counting the number of errors in dialogues. However, we found that most spelling errors affected grammatical words or items not requiring semantic annotation. We thus manually corrected the orthographic errors in the input and evaluated the NLU again. Our results showed that the F-measure did not largely improve (+0.4%). This confirmed that spelling errors had a minor impact on the semantic annotation. Given those results and the space limits of this article, we do not report the evaluation of this module.
5.3.2 Results
We reported a preliminary evaluation based on 242 turn pairs in a previous article (Campillos-Llanos et al. Reference Campillos-Llanos, Bouamor, Zweigenbaum, Rosset and Calzolari2016). Herein, we present the results based upon the data obtained to date from 39 medical users (around 4044 turn pairs). Table 11 shows that the NLU achieves a high F-measure (95.8%), balancing precision (96.8) and recall (94.9). Recall is lower than precision due to missing entity types and unannotated terms. The 6.1% of SER implies that, on average, one entity type was incorrectly labelled every 16.4 labels. The 10.7% of SeER means that, overall, one turn with incorrect labels occurred every 9.3 turns. Both these error rates are low. In terms of impact on dialogue flow, errors at this level made users reformulate their questions or change the topic of the dialogue.
5.4 Quantitative evaluation of dialogue management
5.4.1 Methods
To evaluate the dialogue manager, we adopted the framework explained by Dickerson and colleagues, who evaluated a system developed for the same task and domain (Dickerson et al. Reference Dickerson, Johnsen, Raij, Lok, Hernandez, Stevens and Lind2005). We analysed the logs of user–system interactions and manually classified each turn pair as correct, incorrect, not understood or clarification requests (Purver, Ginzburg and Healey Reference Purver, Ginzburg, Healey, van Kuppevelt and Smith2003). We define a correct reply as that providing both: (1) a coherent answer with regard to the user question and (2) correct information from the VP record. Conversely, incorrect replies are those not succeeding at providing a coherent answer, or those giving erroneous information. Three of these criteria also map to those applied by Traum and his team (Traum, Robinson and Stefan Reference Traum, Robinson and Stefan2004): (1) correct utterances correspond to Traum and colleagues’ Get response and Appropriate continuations; (2) clarification utterances correspond to Request for repair; and (3) incorrect utterances correspond to Inappropriate response or continuation. A computational linguist analysed interaction logs and classified them; then, a subset of 350 (8.6%) turn pairs, which were hard to interpret and classify, was double-checked by a senior computational linguist; finally, a consensus was reached. We computed the percentage of turn–reply pairs for which the annotations made by one researcher were confirmed by the senior researcher at the consensus stage. The agreement between both linguists was of 93.6%.
Table 10. Evaluation metrics for the NLU evaluation
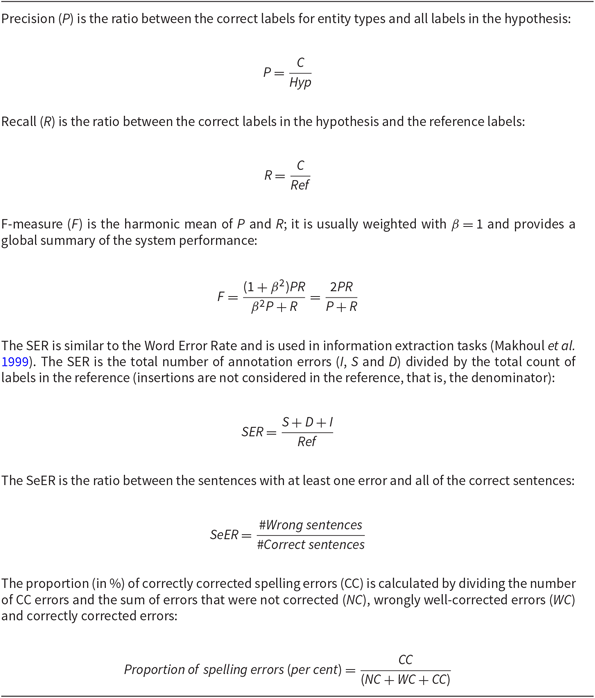
Table 11. Evaluation of the entity detection (NLU)

We report the average number of entities per turn annotated in system hypothesis, standard deviation (stdev), minimum and maximum number of entities. Values for precision, recall, F-measure, SER and SeER are given in percentages, as well as figures between brackets.
5.4.2 Results
An analysis of the user–system interaction logs was performed on the data collected from non-medical users in the development stage and showed that 72.9% of the replies were correct. The analysis of the interaction logs collected from medical users is shown in Figure 9 and Table 12. Segment labeled 1 in Figure 9 stands for correct replies, which represents 74.3%; segment 2, the ratio of incorrect replies (14.9%); segment 3, the proportion of not-understood replies (7.8%); and segment 4, the ratio of clarification requests (2.9%). Performance across VP cases varied (standard deviation, stdev, of 9.5) due to the different number of dialogues conducted with each case, and also in relation to the medical specialities of the cases. We obtained the best results (93.8%) with a VP case suffering from diarrhoea, and poor results (53.6% of correct replies) with a postpartum case from the obstetrics speciality; however, both of these were tested by only one evaluator. In our error analysis of the logs of the postpartum case, we noticed that some of the evaluator’s questions referred to the patient’s newborn. The dialogue manager provided wrong replies, because these question types did not refer to the patient’s medical condition, but to that of her newborn, and the system could not distinguish them. Among incorrect replies, about 37.8% were due to errors in the dialogue manager and 26.2% were caused by unforeseen question types (e.g. we did not prepare rules for questions on the patient’s blood group). Among the not-understood replies, 48.2% were caused by unforeseen question types and about 10.2% were caused by missing variants of questions.

Figure 9. Dialogue manager evaluation.
Table 12. Dialogue manager evaluation (medical doctors)

5.5 Qualitative evaluation of system performance and usability
5.5.1 Methods
Right after users interacted with the system, they filled in a questionnaire with questions using a 5-point Likert scale. The survey addressed the following aspects:
• Global functioning: an overall assessment of system performance.
• Coherence: adequateness of system answers in relation to user input.
• Informativeness: satisfaction with the information provided by the system.
• User understanding: degree of comprehension of system replies by the user.
• System understanding: system’s degree of comprehension of user input.
• Speed: system quickness in replying.
• Tediousness: verbosity of information answered by the system.
• Answer concision: quality of replies in terms of length.
• Naturalness of replies: realism of the utterances produced by the system.
5.5.2 Results
The 131 questionnaires collected from medical users scored highly the degree to which users understood system replies (64.1% of evaluators assessed it very good) and the speed in providing an answer (very good, 69.5%). The following aspects were in general considered good: overall performance (63.4% of users), informativeness (62.6%), coherence of replies (61.1%), system understanding of input (56.5%), concision of replies (45.8%) and their (absence of) verbosity or tediousness (51.9%). The naturalness of replies was scored as good by 45.0% of participants; 29.8% gave a neutral score and 9.9% assessed it as poor. There is still room for improvement for this and other aspects; lower scores, however, represented only a small proportion of users.
Figure 10 depicts the results of the evaluation through the 40 questionnaires collected from participants with computer science backgrounds (left) and the 131 questionnaires filled by medical doctors (right). Assessments were rather similar excepting slight variations regarding informativeness and system understanding (slightly higher for computer science users) or naturalness (slightly higher for medical users). These differences might be due to the more strict criteria applied by medical users and to the improvements made to the system between evaluation rounds.

Figure 10. Qualitative evaluation by non-medical users (left) and medical users (right).
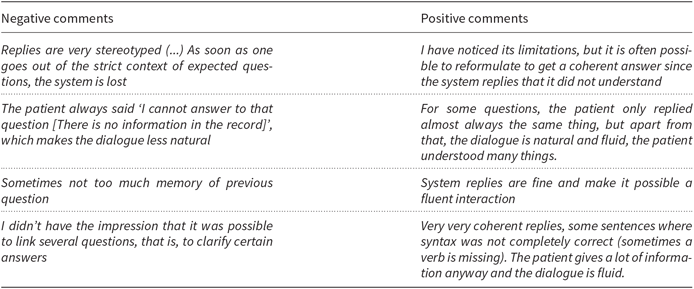
Users provided free comments concerning aspects to be improved. Table 13 shows some of them (translated from French). Several users commented upon difficulties in getting more details after a general question. Sometimes the record lacks detailed information: this raises the question of what the system should answer if the user asks for such missing information. For example, some users asked for the patient’s disease or symptoms, and after the system replied, they wanted to know specific observations, which were not present in the record. In that situation, currently, the dialogue manager gives an explicit answer (I cannot answer that question. This piece of information is not present in the record). This does not always satisfy users due to the missing data or to the lack of naturalness of the reply (see Table 13). Because medical users need accurate information from the patient, we chose to give a neutral reply when no data are available. Likewise, some context processing errors have hindered the correct interpretation of questions. The first case requires to improve the ergonomy of the system; the latter case requires to improve its robustness.
Table 13. A selection of positive and negative user comments in the qualitative evaluation (translated from French)

5.6 Quantitative evaluation of vocabulary coverage
5.6.1 Methods
We assessed how robust our lexicons are by comparing them to domain data not used for developing the system. Because no preexisting library of VP cases existed in French, we used the most similar source we could find. We collected 169 cases from Epreuves Classantes Nationales (‘National Classifying Tests’, hereafter ECN), which are used to prepare exams in medical universities.Footnote l We used the description of the case, not the feedback for students. Table 14 shows a sample.
The procedure was as follows. We lowercased and tokenised the ECN texts; we removed numbers, dates, punctuation and stop words; and we expanded common abbreviations (e.g. mg → milligrams). Then, we compared the word types (i.e. different word forms, not tokens, which represent the occurrence of each type) in these texts against all the terms in the lexical resources used by the NLU, entity normalisation and generation steps. We computed for each text the proportion of in-vocabulary and OOV words, and their average over all texts.
We also evaluated to what extent the ECN texts are different to the first cases used in system development. For this purpose, we compared the word types in the ECN cases to the word types in the initial cases and computed the percentage of ECN words that were not present in the development cases.
Table 14. Sample case of the Epreuves Classantes Nationales (ECN, ‘National Classifying Examination’), and its translation

ECN were used for evaluating the vocabulary coverage of the system with regard to new clinical cases.
5.6.2 Results
Cases in our development set included 1504 tokens (428 types), and two cases had several consultations. We evaluated the system’s vocabulary coverage of the 169 new cases found in ECN texts (24,521 tokens, 4112 types). These counts do not include dates and numbers, which we removed from these texts, because they are not managed through dictionaries and hence raise no coverage problem. We measured that 3805 (92.5%) of the word types occurring in the ECN texts did not occur in the development cases. This shows that the ECN texts used to test vocabulary coverage are really different from the cases used in system development.
The system’s resources recognised an average of 97.85% of ECN word types (stdev = 1.91). A very low percentage of ECN word types were OOV items (2.15%), with an average of 2.34 per case (stdev = 2.72).
Figure 11 shows that missing terms were mostly acronyms and abbreviations (e.g. rcp), domain terms (e.g. decubitus) and medications (e.g. zopiclone). Other OOV words were names of people or locations (e.g. France), other words (e.g. rendez-vous) and spelling errors (*oédème instead of oedème, ‘edema’). Note that names of people and locations are not useful in the system and are thus not included in our lexicons. Accordingly, the actual rate of OOV words is even lower than measured: resources covered an average of 98.09% of ECN word types (stdev = 1.86), and the average of OOVs per case in the ECNs was of 2.14 (stdev = 2.67).

Figure 11. Breakdown of types of OOV words in ECN texts.
5.7 Qualitative analysis of vocabulary usage
5.7.1 Methods
To illustrate the difficulties in managing the variety of terms in our task, we provide a qualitative analysis of domain term usage in the interaction data. First, we analysed how medical evaluators used domain terms in dialogue logs. We only analysed dialogues in cases tested by more than one medical doctor (28 different cases). As an illustration, we focused on terms related to entity types of specific references to diseases (disease_spec) and symptoms (symptom_vb, which labels verb forms, and symptom). We examine here the actual occurrence of the terms observed in the interaction logs. Second, we analysed the term distribution in the corresponding VP records (only for those 28 cases). We did not include stop words nor the record section containing demographic data (e.g. proper names or civil status). For both aspects of the analysis, we obtained frequencies of usage of each item and plotted them to examine their distribution across user interactions or records.
5.7.2 Results
In the interaction logs and patient records of the 28 analysed cases, we found 1408 different tokens. Less than 60 (∼4%) have a frequency over 10 and around 30 (∼2%) occur in at least 10 cases. Figure 12 summarises our results. The frequency distribution of the terms for symptoms and diseases are represented in two plots (A and B). Terms were analysed in the dialogues of all VP records (plot A) and we also counted the number of different cases where each term occurred in the interaction dialogues (plot B). Secondly, we plot the analysis of tokens in all records: token frequency in all records (plot C) and the number of cases where each token appeared (plot D). All plots show that both token frequency and terms of these entity types follow a Zipfian-style distribution. Term management in our system needs to cope with a distribution of Large Number of Rare Events.

Figure 12. Qualitative analysis of term distribution in dialogues and VP records.
Table 15. Most and least frequent terms (translated from French) observed in dialogues with 28 different VPs for entity types of symptoms and diseases

Note that some terms cannot always be assigned to symptom or disease, and the dialogue context or VP record are needed to make a distinction. For example, vertigo is most commonly a symptom, but it might be a chronic condition in a specific case.
Table 16. Most and least frequent tokens (translated from French) observed in the 28 analysed VP records (only a selection of the least frequent tokens is shown)

A quick look showed that very low-frequency items are domain terms (e.g. cianotic or acetylleucine). This illustrates the difficulty of term detection in our task and justifies our approach based on rich terminological resources. The methods we propose enhance the system’s ability to adapt to different cases and detect rare and unseen vocabulary items for a successful interaction. Table 15 breaks down the most frequent terms for specific references to symptoms (labels symptom and symptom_vb) and diseases (disease_spec) observed in the dialogues, and Table 16 reports the frequency analysis of tokens in the records.
5.8 Discussion
Our system is aimed at dealing with new cases and therapeutic areas. To achieve that, we rely on robust and comprehensive terminology components, which, as far as we know, are unparalleled in current VP systems (Maicher et al. Reference Maicher, Danforth, Price, Zimmerman, Wilcox, Liston, Cronau, Belknap, Ledford, Way, Post, Macerollo and Rizer2017). The method which consists in crowd-sourcing the evaluation of the answers of the dialogue system and the extending its question–answer database with the correct answer (Rossen, Lind and Lok Reference Rossen, Lind, Lok and Ruttkay2009) does not seem scalable for extending the components of our system and its terminology. Because these variation phenomena are easier to process through thesauri, we rely on domain lexicons and ontological knowledge. Our approach is closer to that based on a taxonomy of questions (Talbot et al. Reference Talbot, Kalisch, Christoffersen, Lucas and Forbell2016); however, we use terminologies available in the UMLS Metathesaurus.
Evaluation outcomes showed that the core lexical and terminological components seem stable and able to process new clinical cases. From a quantitative point of view, the NLU module achieved an F-measure of 95.8%, balancing precision (96.8%) and recall (94.9%) when annotating entities in user input.
The test of vocabulary coverage brought out the few types of terms that occurred in unseen patient cases and were still missing in our lexicon: these only represented 2.16% of terms in a collection of 169 descriptions of clinical cases, and most missing terms were acronyms and abbreviations. From a qualitative point of view, users who tested the system did not mention any error related to terminology needs.
According to the evaluations, the most important causes of failures in the dialogue manager are beyond terminology needs and might define the limits of a purely rule- and frame-based system. The difficulty of the task also accounts for processing failures of follow-up queries after general questions, especially in cases of missing information in the record. Medical doctors tend to ask general questions to begin to circumscribe a diagnosis; then, if the patient replies with the searched bit of information, they ask for more details. This requires both processing correctly the implicit information in the dialogue context and foreseeing all details to be queried, for example, observations, descriptions (e.g. intensity) or temporal data related to a condition. The lack of pre-existing task-specific dialogue data hinders achieving a comprehensive coverage of question types, query variants and interaction contexts.
We would like to improve the naturalness of replies, especially those with long sentences and negative symptoms. The realism of responses depends on technical aspects as well as on how medical instructors input data.
The methods we propose should be valid for other dialogue tasks in other domains where rich lexical and/or ontological resources exist and a semi-structured database is available. That makes it possible to develop a system to collect interaction data, which can then be used in statistical or machine-learning approaches. Nonetheless, a key takeaway is the fact that, even when rich resources exist, these need an extensive effort of iterative filtering and task adaptation before production mode. As we explained, we created ad hoc lexicons with lay variants or equivalences between noun terms and verbs (which are missing in the UMLS). Similar needs were reported when adapting the UMLS for concept indexing (Nadkarni, Chen and Brandt Reference Nadkarni, Chen and Brandt2001).
In terms of dialogue management, we contribute with an approach for querying the database at each dialogue move by considering the semantic content of each dialogue state.
6. Conclusion
In this paper we highlighted the difficulties raised by the terminological needs of a dialogue system that aims at providing natural language interaction in a simulated medical consultation context, robust enough for multiple clinical cases. We designed three models involved in such a dialogue task: a patient record model, the knowledge model for the task and a termino-ontological model.
This work focused on the termino-ontological model, which manages terminological and linguistic resources. To populate the model, we adopted a comprehensive approach to lexicon and terminology collection. We collected term variants for domain concepts based on existing medical terminologies, which helped us structuring terms according to the concepts they describe. We compiled large dictionaries of inflectional and derivational word variants. These resources enabled the system to: (1) recognise a large number of entities in the NLU step; (2) handle general and specific entities in the NLU or dialogue manager modules; (3) perform entity linking, entity normalisation and hierarchical reasoning; and (4) give priority to lay variants in the generation step. The quality of the collected resources allowed the system to obtain a high vocabulary coverage when tested on a large number of unseen cases: the system proved stable for the task and robust enough to cope with the vocabulary of new cases. Our system stands out from current research on VPs by its ability to handle a large variety of clinical specialities and cases. We developed the system with 35 different records from 18 medical specialities.
A total of 32 non-medical users and 39 medical students and doctors evaluated the system. Overall, the majority of users evaluated it as good or very good in most dimensions. The evaluation also highlighted aspects that deserve further work. User comments in the evaluation reflected these shortcomings, especially regarding follow-up utterances after general questions, improving the naturalness of some replies and handling missing information in the patient record.
We make the evaluation corpus used in this work available for the community.Footnote m In the context of a lack of dialogue resources, especially in the medical domain, we believe these data will be useful for moving ahead in the field.
Now that we have collected interaction corpora; we are focusing our research on machine-learning-based methods. Specifically, we have begun exploring the classification of question types according to the system needs, that is, the system’s current rules or an alternative processing strategy (Campillos-Llanos, Rosset and Zweigenbaum Reference Campillos-Llanos, Rosset and Zweigenbaum2017). Designing such a fallback strategy is our research interest, with a view to providing a satisfactory answer when a question cannot be handled with the current approach. We estimate that a subset of system’s wrong and not-understood replies (overall, less than 20% of replies) would need a fallback strategy.
The system has been adapted to English and Spanish, following the same design procedures and models (e.g. entity types scheme) as explained. Domain lists contain over 116,000 terms in English and 103,000 in Spanish, and dictionaries gather over 1,886,000 word/concept entries in English and 1,428,000 in Spanish. A thorough data collection and evaluation are needed to improve these versions of the system.Footnote n
Author ORCID
Leonardo Campillos-Llanos 0000-0003-3040-1756
Acknowledgements
This work was funded by BPI through the FUI Project PatientGenesys (F1310002-P) and by SATT Paris Saclay through project PVDial. We greatly thank all the evaluators who tested and assessed the system, and we thank VIDAL®, who provided a domain corpus of medications and disorders. We also thank Brigitte Grau and the anonymous reviewers for their valuable remarks on the manuscript.