Introduction
The genus Sinistroporomonorchis Wee, Cutmore, Pérez-del-Olmo & Cribb, 2020 is a small group of trematodes belonging to the Monorchiidae Odhner, 1911 with 5 species described thus far (Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023). This genus was proposed by Wee et al. (Reference Wee, Cutmore, Pérez-del-Olmo and Cribb2020a) to include 2 species, Sinistroporomonorchis glebulentus (Overstreet, Reference Overstreet1971) and S. lizae (Liu, Reference Liu2002); both species are parasites of mugilids. The distinguishing characters that separate the genus from most other monorchiids is the possession of a distinctly sinistral genital pore, an unspecialized oral sucker, short oesophagus and restricted vitelline masses (Wee et al., Reference Wee, Cutmore, Pérez-del-Olmo and Cribb2020a). More recently, Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023) performed morphological and molecular analyses including the 28S region of the rDNA and cox1 of the mitochondrial DNA to describe 3 species of Sinistroporomonorchis from mugilids in the Yucatán Peninsula, Mexico, namely S. mexicanus (Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023); S. yucatanensis (Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023) and S. minutus (Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023).
Four of the five species were described from Mugil spp. (Mugilidae) in the Americas whereas S. lizae was described from Planiliza carinata (Valenciennes) (Mugilidae) in Asia (Overstreet, Reference Overstreet1971; Liu, Reference Liu2002; Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023). However, Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023) reported that Lasiotocus sp. ex Menidia menidia (L.) (Atherinopsidae) from New Jersey, USA sequenced by Panyi et al. (Reference Panyi, Curran and Overstreet2020), should be considered as a member of Sinistroporomonorchis based on results of the 28S analysis (see Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023). Additionally, Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Guillén-Hernández, Badillo-Alemán, Chiappa-Carrara and Pérez-ponce de León2023, Reference Espínola-Novelo, Solórzano-García, Badillo-Alemán, Chiappa-Carrara, Guillén-Hernández and Pérez-ponce de León2025) reported Sinistroporomonorchis sp. based on the sequence data of 28S rDNA of one individual obtained from Floridichthys polyommus (Hubbs) and one from Fundulus persimilis (Miller) in La Carbonera coastal lagoon, Yucatán, Mexico.
In the present study, we are completing the inventory of the parasite fauna of the F. polyommus; newly collected individuals of trematodes consistent with the concept of Sinistroporomonorchis were obtained from F. polyommus in 3 coastal lagoons and 1 locality off the coast of the Yucatán Peninsula. A detailed morphological examination includinga principal component analysis (PCA) of those specimens as well as scanning electron microscopy (SEM) photomicrographs, in combination with information from nuclear and mitochondrial DNA suggested that these specimens represented a new species of Sinistroporomonorchis. The new species is described herein.
Materials and methods
Sample collection
A total of 119 individuals from F. polyommus and 41 individuals from F. persimilis were collected using cast nets from 3 coastal lagoons and 1 locality off the coast the Yucatán Peninsula (YP) between 2023 and 2024 (Table 1; Figure 1). Fishes were kept alive and examined for helminths a few hours after capture. Individual fish was euthanized following the procedures accepted by the American Veterinary Medical Association (AVMA, 2020), dissected and immediately examined under a stereomicroscope. Sixty-two individuals of Sinistroporomonorchis were recovered from the intestines of 28 specimens of F. polyommus, and only 1 individual was recovered from an individual of F. persimilis (Espínola-Novelo et al., Reference Espínola-Novelo, Solórzano-García, Badillo-Alemán, Chiappa-Carrara, Guillén-Hernández and Pérez-ponce de León2025) (Table 1). Most of the sampled specimens of Sinistroporomonorchis were juveniles (56%). Monorchiids were fixed in hot distilled water and preserved in 100% ethanol for morphological and molecular analysis.

Figure 1. Sampling localities within the coastal lagoons of the Yucatán Peninsula, Mexico. Localities correspond with Table 1.
Table 1. Locality, collection date, host species, host length, prevalence, intensity of infection and GenBank accession numbers of specimens collected in this study

TL, mean total length of hosts; HI/HR, host infected/host revised; IoI, intensity of infection.
Numbers for localities correspond to Figure 1.
* Published in Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Badillo-Alemán, Chiappa-Carrara, Guillén-Hernández and Pérez-ponce de León2025).
Morphological analyses
Some specimens were post-fixed in hot formalin to harden the tegument. Posteriorly, those specimens were dehydrated through a graded alcohol series, stained with Mayer’s paracarmine (Merck, Darmstadt, Germany), cleared with methyl salicylate and mounted on microscope slides with Canada balsam. Mounted specimens were photographed, examined morphologically and measurements were obtained using under a bright field Nikon DS-Ri1 microscope with Nikon NIS Elements microscope software (Nikon). Measurements are given in micrometres (μm). Drawings were made with Adobe Illustrator 25.4.1 (Adobe, Inc.). Holotype and paratypes were deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City.
For SEM, specimens were dehydrated in a graded ethanol series, critical point dried and sputter coated with gold. SEM images from the monorchiids were obtained with a Hitachi Stereoscan Model SU1510 SEM at 15 kV at the Laboratorio de Microscopia y Fotografía de la Biodiversidad, Instituto de Biología, Universidad Nacional Autónoma de México.
A PCA was implemented to explore and describe the patterns of morphological variation of 7 specimens of Sinistroporomonorchis collected from F. polyommus. Morphometrical data of the 23 specimens reported by Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023) belonging to 4 species, Sinistroporomonorchis glebulentus, S. mexicanus, S. minutus and S. yucatanensis (CNHE 11838–11844) were used to complement this analysis (Supplementary Table S1). The species S. lizae was not included in the analysis since we have no access to the original measurements, necessary to run the analysis. The PCA was conducted using ggplot2, ggfortify, cluster and lfda implemented in R (R Core Team, 2020) and included 58 metrical data (see Supplementary Table S1).
Amplification and sequencing of DNA
Six specimens of Sinistroporomonorchis sp. collected from F. polyommus were placed individually in tubes with a digestion solution for DNA extraction at 56°C overnight. The digestion procedure, amplification and sequencing protocols followed Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023). The domains D1–D3 of the large subunit of nuclear ribosomal RNA gene (28S) were amplified via PCR using the primers: 391 5ʹ-AGCGGAGGAAAAGAAACTAA-3ʹ and 536: 5ʹ-CAGCTATCCTGAGG GAAAC-3ʹ for 28S (García-Varela and Nadler, Reference García-Varela and Nadler2005). Additionally, a fragment of cox1 was amplified for 2 specimens. In addition, the individual of Sinistroporomonorchis sp. sequenced by Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Guillén-Hernández, Badillo-Alemán, Chiappa-Carrara and Pérez-ponce de León2023) for 28S (OR537912) was sequenced for cox1 in the present study. For amplifying the cox1 region, the primers designed by McNamara et al. (Reference McNamara, Miller and Cribb2014) were used, i.e. cox1tremF (5ʹ-TTCACKTTGGATCATAAGCGT-3ʹ) and Mon.mt3 (5ʹ-ACCATAAACATRTGRTG-3ʹ). Sequences were assembled and edited using Geneious v7 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012) and deposited in the GenBank database.
Alignments and phylogenetic analyses
Sequences obtained in the current research from 28S rDNA and cox1 mtDNA were aligned separately with data from 5 species of Sinistroporomonorchis plus 3 unidentified sequences of Sinistroporomonorchis, 1 from Menidia menidia (MN984477), 1 from F. polyommus (OR537912) and 1 from F. persimilis (PP919655), all downloaded from the GenBank dataset. Three species of Allobacciger Hafeezullah & Siddiqi, 1970, one of Alloinfundiburictus Wee, Cutmore, Pérez-del-Olmo & Cribb, 2020 and one of Monorchis Monticelli, 1893 were used as outgroups (see Table 2). The alignments were trimmed to the shortest. The 28S rDNA alignment consisted of 46 sequences with 1186 nucleotides and 19 sequences with 496 nucleotides for cox1. Alignments were constructed using the software Clustal W (Thompson et al., Reference Thompson, Gibson, Plewniak and Jeanmougin1997) with default parameters and adjusted manually with the Mesquite software (Maddison and Maddison, Reference Maddison and Maddison2011).
Table 2. Sequences from GenBank used for phylogenetic analysis in the present study

The phylogenetic analyses were performed using Bayesian inference (BI) and maximum likelihood (ML) methods. The BI analysis was inferred with MrBayes version 3.2.7 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2012) through the interface: Cyberinfrastructure for Phylogenetic Research Science Gateway v3.3 (CIPRES) (Miller et al., Reference Miller, Pfeiffer and Schwartz2010) and the ML analysis were carried out with the RAxML version 7.0.4 (Silvestro and Michalak, Reference Silvestro and Michalak2011). The best model was estimated with the Akaike information criterion using the jModelTest version 0.1.1 program (Posada, Reference Posada2008). The best model for each dataset was GTR + I + G. The BI analyses included 2 simultaneous runs of Markov chain Monte Carlo for 10 million generations, sampling every 1000 generations, a heating parameter value of 0.2 and a ‘burn-in’ of 25%. To support each node in the ML analyses, 10 000 bootstrap replicates were run. Trees were drawn using FigTree program v.1.3.1 (Rambaut, Reference Rambaut2012). The genetic divergence among taxa was estimated using uncorrected ‘p’ distances in MEGA v. 6. (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013) (see Table 3).
Table 3. Pairwise nucleotide sequence comparisons between taxa for the aligned 28S rDNA sequences (1186 nt) below the diagonal and for cox1 sequences (496 nt) above the diagonal

Bold represented the genetic intraspecific divergence.
Results
Taxonomy
Platyhelminthes Minot, 1876
Trematoda Rudolphi, 1808
Plagiorchiida La Rue, 1957
Monorchiidae Odhner, 1911
Sinistroporomonorchis Wee, Cutmore, Pérez-del-Olmo & Cribb, 2020
Sinistroporomonorchis bolini n. sp. (Figure 2)

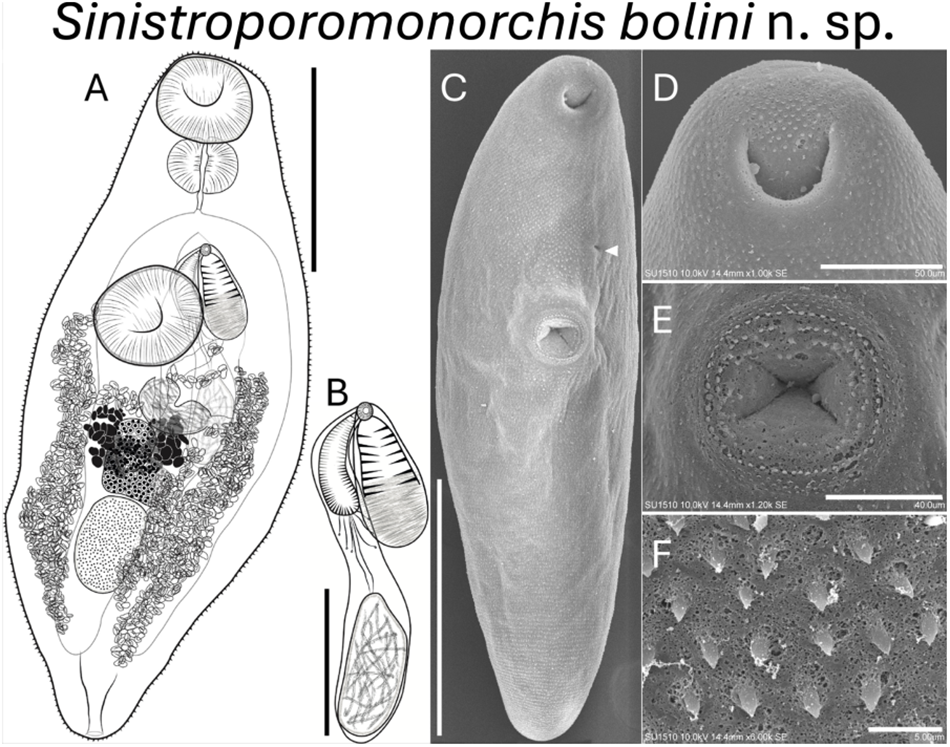
Figure 2. Sinistroporomonorchis bolini n. sp. from Floridichthys polyommus. (A) Whole worm holotype, ventral view; (B) terminal genitalia of holotype; scanning electron micrographs of voucher; (C) whole worm; (D) oral sucker; (E) ventral sucker; (F) tegumental spines. Scale bars (μm) = (A) 200; (B) 100; (C) 300; (D) 50; (E) 40; (F) 5. Arrow marks genital pore.
Type host: Floridichthys polyommus Hubbs, Ocellated killifish, Cyprinodontidae
Additional host: Fundulus persimilis Miller, Yucatán Killifish, Fundulidae
Site of infection: Intestine
Type locality: Dzilam de Bravo, Yucatán (21°22ʹ31ʺN; 88°59ʹ44ʺW)
Additional localities: La Carbonera (21°13ʹ54ʺN; 89°53ʹ25ʺW) and San Felipe (21°34ʹ06ʺN; 88°14ʹ13ʺW) lagoons, Yucatán; Off the coast of Xcalak (18°14ʹ32ʺN; 87°53ʹ06ʺW), Quintana Roo.
Specimens deposited: 1 Holotype (CNHE-12163); 6 paratypes (CNHE-12164)
Etymology: The specific epithet bolini refers to the vernacular name of the host, Floridichthys polyommus which is locally known as ‘bolin yucateco’. We treat the word ‘bolin’ as a masculine proper name.
GenBank Accession: PQ634759–PQ634764 for 28S; PQ630635–PQ630637 for cox1
ZooBank LSID: 0A32C2A1-8B29-4D7C-A9C5-B57263159BDA
Sinistroporomonorchis bolini n. sp. (Figure 2).
Description
Based on 7 mature individuals (Range followed by the mean in parentheses). Body fusiform 561–742 (675) long, 175–288 (216) wide, widest at midbody; 2.4–3.7 (3.1) times longer than wide. Tegument thin, armed with small spines throughout body. Forebody relatively short 209–262 (226) long, occupies 29.6–37.6 (33.7)% of body length. Hindbody 282–424 (354) long, occupies 49.7–57.1 (52.4)% of body length. Oral sucker spherical, terminal, with ventral aperture, 56–91 (76) long, 57–101 (83) wide. Ventral sucker round located anterior to middle of body, 65–128 (91) long, 68–108 (83) wide. Oral sucker length to ventral sucker length ratio 1: 0.64–1.01 (0.84). Oral sucker width to ventral sucker width ratio 1: 0.83–1.13 (0.97). Prepharynx observed in some specimens, 2–8 (5) long. Pharynx well-developed, spherical, large, 41–59 (51) long, 57–84 (71) wide; 58.8–88.5 (68.3)% of oral sucker length; 75.2–100 (87.3)% of oral sucker width. Oesophagus short 26–90 (54) long, occupies 3.6–12.5 (7.9)% of body length. Intestinal bifurcation in forebody, well anterior to ventral sucker; pre-bifurcal zone 100–221 (162) long, occupies 13.4–30.7 (24)% of body length. Intestinal caeca robust, long, terminate posterior to testis. Post-testicular zone 34–161 (102) long, or 6–22.5 (14.9)% of body length from posterior end of body.
Testis single, ovoid, 80–126 (105) long, 52–81 (62) wide; posterior to midbody, 14.3–20.7 (17.9)% of body length from ventral sucker, 84–154 (121). Pre-testicular zone 365–480 (433) long, occupies 59.7–66.8 (64.2)% of body length; post-testicular zone 109–166 (136) long, or 16.8–23.8 (20.2)% of body length. Cirrus-sac arcuate, dorsal to ventral sucker extending to middle of body, mostly intercaecal, slightly overlaps left caecum, 169–243 (203) long, 27–45 (34) wide, occupies 26.1–34.3 (30.6)% of body length. Seminal vesicle elongate, unipartite, 25–101 (54) long, 22–46 (37) wide; occupies 12.4–41.5 (26.6)% of cirrus-sac length. Pars prostatica simple, with few prostatic cells observed, 24–37 (33) long. Cirrus thick, armed with spines, 74–145 (108) long, 15–32 (20) wide, occupies 40.3–77.1 (53.6)% of cirrus-sac length. Genital atrium unspined. Common genital pore small, sinistral to midline, anterior to ventral sucker, located close to intestinal bifurcation.
Ovary lobulate, slightly irregular and elongate transversally, in anterior half of hindbody, distinctly posterior to ventral sucker, overlaps right caecum ventrally, 40–89 (67) long, 45–78 (57) wide; pre-ovarian zone 145–421 (341) long, or 24.8–58.6 (50.8)% of body length; post-ovarian zone 138–289 (225) long, or 23.7–41.5 (33.7)% of body length. Vitellarium composed of 2 dense clusters of follicles that are confluent at the posterior half of the clusters, irregularly shaped, distributed closely and dorsal to ovary, 45–78 (63) long or 7.7–10.9 (9.3)% of body length. Uterus mostly restricted to hindbody in lateral fields of the body, thin-walled, extensive, ventral to ovary, testis, caeca and part of cirrus-sac, with coils mostly indiscernible; ascending coil forms metraterm and enters terminal organ at posterior end. Terminal organ sinistro-ventral to cirrus-sac, 86–120 (100) long, 41–78 (53) wide, comprising unspined posterior chamber, and spined anterior section. Posterior chamber spherical, containing fibrous mass 26–51 (36) long, 33–55 (44) wide. Anterior section armed with long and thin spines, 47–85 (63) long, 24–61 (61) wide. Seminal receptacle canalicular, anterior to ovary. Eggs slightly tanned, operculate, unfilamented, 7–19 (12) long, 3–9 (7) wide. Excretory vesicle Y-shaped. Excretory pore terminal.
Morphometric analyses
PCA was conducted to corroborate the morphological differences between the new species and those previously described by Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023). A total of 58 variables were considered from 30 specimens corresponding to 5 species, S. bolini n. sp., S. glebulentus, S. minutus, S. mexicanus and S. yucatanensis. The morphometrical data of the new species from F. polyommus formed a separate cluster from the other species, particularly along PC2, corroborating the separation of this species from the other congeners. In addition, S. minutus was also separated, as well as the 2 individuals of S. glebulentus. Only 1 individual from S. mexicanus was grouped with S. yucatanensis (Figure 3).

Figure 3. Principal component analysis of 5 species of Sinistroporomonorchis conducted with 58 morphometric variables from 30 individuals.
Remarks
The new species, S. bolini n. sp. collected from F. polyommus in 4 localities of the Yucatán Peninsula, Mexico is consistent with the generic concept of Sinistroporomonorchis in the possession of a sinistral genital pore, an unspecialized oral sucker, and restricted vitelline masses. With the addition of the new species, the genus Sinistroporomonorchis now contains 6 species. The newly described species overlaps in most of the metrical characters in comparison with the remaining 5 species. However, S. bolini n. sp. can be differentiated from the 5 Sinistroporomonorchis species by the presence of an overall larger pharynx; in the new species pharynx is 41–59 (51) long and 57–84 (71) wide, whereas the range of the other 5 species varies between 22 and 46 long and 30–55 wide. Likewise, the proportion of pharynx width to oral sucker width could also be a reliable character to distinguish the new species, 0.75–1 (0.87); meanwhile, the range of the other 5 species is 0.41–0.84. Additionally, S. bolini n. sp. can be further differentiated from the other congeners by possessing a uterus arranged in 2 main lateral fields overlapping the caeca; in addition, caeca are very wide (robust); all other Sinistroporomonorchis spp. possess simple caeca and the uterus is not distributed in 2 lateral fields. Furthermore, in the PCA, the individuals of the new species formed a separated cluster from the other species of Sinistroporomonorchis. This result supports the validity of the new species.
Molecular data and phylogenetic analysis
28S
The phylogenetic analyses inferred with ML and BI recovered similar topologies (Figure 4A). The genus Sinistroporomonorchis was yielded as monophyletic, with high nodal support although it formed an unresolved clade with Monorchis and Alloinfundiburictus with strong support (1/100). Within the genus Sinistroporomonorchis, 2 subclades were formed. The first major subclade contained Sinistroporomonorchis sp. (MN984477), as the sister taxa of 2 groups, one containing S. lizae, and S. glebulentus + S. mexicanus, and the other containing the isolates of S. bolini n. sp. with high to moderate nodal support (1/86). The second major subclade was formed by Sinistroporomonorchis minutus + S. yucatanensis, with strong support (1/100). The 6 new sequences of S. bolini n. sp. from F. polyommus, plus the 2 sequences (OR537912, PP919655) obtained by Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Guillén-Hernández, Badillo-Alemán, Chiappa-Carrara and Pérez-ponce de León2023); Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Badillo-Alemán, Chiappa-Carrara, Guillén-Hernández and Pérez-ponce de León2025) were nested within a clade with strong support (1/100) corroborating the conspecifity, although some isolates grouped in 2 separate groups, but with very low sequenced divergence. The new species was recovered as the sister taxa of the clade formed by S. lizae, and S. glebulentus + S. mexicanus with strong to moderate nodal support (0.98/68).

Figure 4. Phylogenetic trees inferred from the Bayesian inference and maximum likelihood analyses of the (A) 28S gene and (B) cox1 gene. Posterior probabilities (BI) and bootstrap values (ML) are shown above and below the nodes, respectively.
The genetic divergence estimated for 28S rDNA between the new species and all other representative species of Sinistroporomonorchis ranged from 4.1 to 9.1%. The genetic divergence between the new species and the closely related species (S. lizae, S. glebulentus and S. mexicanus) ranged from 4.6 to 7.1%. The genetic divergence between the new species and the second subclade (Sinistroporomonorchis minutus + S. yucatanensis) is even higher, varying 6.4–9.1%. The intraspecific genetic divergence among the 8 new sequences of S. bolini n. sp. varied between 0 and 1.4% (Table 3).
cox1
The phylogenetic analyses inferred with ML and BI also recovered similar topologies (Figure 4B), although all branches were resolved. The analyses also showed Sinistroporomonorchis resolved as monophyletic with high nodal support (0.99/97). Two subclades were also formed, however, unlike 28S analysis, the phylogenetic position of the new species was different. In the first subclade, S. glebulentus was yielded as the sister species of S. mexicanus. In the second subclade, the new species was nested as the sister taxa of S. minutus + S. yucatanensis albeit with very low support values (0.59/59). The 3 newly sequenced individuals of S. bolini n. sp. formed a separated clade with strong nodal support (1/100).
The genetic divergence estimated for cox1 between the new species and its sister taxa (S. minutus + S. yucatanensis) was 18.3–22%. The divergence between the new species and the other major subclade (S. glebulentus + S. mexicanus) was 17.7–20%. Finally, the intraspecific genetic variation among the 3 newly sequences of S. bolini n. sp. was 0.8–11.4% (Table 3).
Discussion
Members of Sinistroporomonorchis are typically associated with mugilid hosts (Mugilidae). However, Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023) concluded that Lasiotocus sp. reported by Panyi et al. (Reference Panyi, Curran and Overstreet2020) from the atherinopsid Menidia menidia should also be considered as a member of Sinistroporomonorchis. Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Guillén-Hernández, Badillo-Alemán, Chiappa-Carrara and Pérez-ponce de León2023) and Espínola-Novelo et al. (Reference Espínola-Novelo, Solórzano-García, Badillo-Alemán, Chiappa-Carrara, Guillén-Hernández and Pérez-ponce de León2025) then reported the presence of Sinistroporomonorchis sp. also from 2 non-mugilid species, the cyprinodontid F. polyommus and the fundulid F. persimilis in La Carbonera coastal lagoon, Yucatán Peninsula, Mexico but were unable to identify the species due to the limited number of specimens obtained from these hosts. This indicates that the genus can no longer be considered specific to mugilids, since apparently several host-switching events have occurred during the evolutionary history of the group.
In this study, following an integrative taxonomy approach, we describe Sinistroporomonorchis bolini n. sp. from F. polyommus and F. persimilis in 4 localities of the Yucatán Peninsula, Mexico. The new species is the sixth described for the genus Sinistroporomonorchis and the fifth described from the Yucatán Peninsula. It is worth noting that prevalence and abundance values of S. bolini n. sp. is relatively low compared with values of these ecological parameters obtained for other Sinistroporomonorchis species (see Overstreet, Reference Overstreet1971; Liu, Reference Liu2002; Andrade-Gómez et al., Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023); this may indicate that in fact mugilids are the main host group, and that the presence of species of Sinistroporomonorchis in other fish groups indicate a recent diversification. Also, only 1 individual of the new species was found in 41 sampled specimens of F. persimilis studied in 2 localities of the Yucatán Peninsula, suggesting this may be an accidental infection. Furthermore, most of the specimens of S. bolini n. sp. found in F. polyommus were juveniles which may indicate the novel development of the new species in a non-mugilid host that occur in sympatry with mugilids and is apparently experiencing an incipient diversification process. However, it is clear that the high genetic divergence observed between the new species and other congeners for both molecular markers confirmed the validity of the new species.
In this study, we recovered the same topology as in Andrade-Gómez et al. (Reference Andrade-Gómez, Ortega-Olivares, Solórzano-García, García-Varela, Mendoza-Garfias and Pérez-ponce de León2023) (Figure 4). However, the phylogenetic position of the new species with respect to the other congeners differed with each molecular marker. In the 28S analysis, S. bolini n. sp. was placed in the first subclade as sister group to a clade formed by S. glebulentus, S. mexicanus and S. lizae. Meanwhile, with cox1 analysis, S. bolini n. sp. was recovered in the second major subclade as the sister group of S. minutus and S. yucatanensis, although with weak nodal support. Based on these results, we consider that the 28S analysis provide, as in other trematodes, a better understanding of the evolutionary interrelationships among Sinistroporomonorchis as discussed by Pérez-Ponce de León and Hernández-Mena (Reference Pérez-Ponce de León and Hernández-Mena2019).
Finally, this study represents an effort to continue the description of the diversity of parasites associated with F. polyommus. Still, more information is required to solve the phylogenetic position of the new species, and the evolutionary relationships of Sinistroporomonorchis species and most likely more species of this genus remain undiscovered since they might be associated to a variety of hosts, including species of atherinopsids, cyprinodontids and fundulids.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182025000010.
Acknowledgements
JEN thanks CONAHCYT for the postdoctoral fellowship ‘Estancias Posdoctorales por México, Modalidad Académica’ (CVU 351170). We sincerely thank Maribel Badillo Alemán and Alfredo Gallardo Torres, Laboratorio de Biología de la Conservación, Facultad de Ciencias UNAM for lab facilities and the identification of hosts. We are truly grateful to Berenit Mendoza Garfias, LaNaBio for the help to obtain the SEM images. We are grateful to Laura Marquez and Nelly López, LaNaBio for their help for sequencing DNA. We thank Luis García Prieto for providing access numbers of CNHE. We thank to Norberto Colín for the use of the facility of the Molecular Biology lab at ENES-Mérida. We appreciate the help of Atl Gerardo Palacios Díaz regarding PCA. We also thank Betzi Pérez, José Sandoval, Edith Gámez and Marco Escalante for their help during fieldwork.
Author contributions
LAG and GPPL conceived, designed and wrote the article. LAG, JEN and BSG conducted data gathering. LAG and JEN performed morphological analyses. LAG and BSG performed molecular analysis.
Financial support
This research was supported by grant from the Programa de Apoyo a Proyectos de Investigación e Inovación Tecnológica (PAPIIT-UNAM IN212621) to GPPL.
Competing interests
The authors declare there are no conflicts of interest.
Ethical standards
Specimens were sampled under the collecting permit issued by the Comisión Nacional de Acuacultura y Pesca to Alfredo Gallardo Torres (No. PPF/DGOPA-001/20). Fish were humanely euthanized following the protocols described by the 2020 edition of the AVMA Guidelines for the Euthanasia of Animals.