Introduction
Transgenic cotton expressing Bacillus thuringiensis (Bt) endotoxin Cry1Ac has been widely adopted in the United States, China, India, Australia and South Africa (James, Reference James2006). The major lepidopteran pests targeted by transgenic Bt cotton, such as Helicoverpa armigera, Helicoverpa zea, Heliothis virescens and Petinophora gossypiella, have been selected for high levels of resistance to Bt toxin Cry1Ac in the laboratory (Gould et al., Reference Gould, Anderson, Bumgarner and Moar1995; Liu et al., Reference Liu, Tabashnik, Meyer, Carrière and Bartlett2001; Xu et al., Reference Xu, Yu and Wu2005; Anilkumar et al., Reference Anilkumar, Rodrigo-Simón, Ferré, Pusztai-Carey, Sivasupramaniam and Moar2008). Although resistance to transgenic Bt cotton has not yet evolved in field populations of the target pests, it remains the major threat to the long-term efficacy of transgenic Bt cotton.
It is critical to understand the molecular mechanisms of insect Bt resistance if informed and proactive resistance management tactics are to be developed. Although field populations of cotton pests with Bt resistance are not available, laboratory-selected Bt-resistant insect strains allow us to dissect possible resistance pathways. In three lepidopteran cotton pests, H. virescens, P. gossypiella and H. armigera, mutations in a cadherin gene are genetically linked to Cry1Ac resistance in the laboratory-selected strains (Gahan et al., Reference Gahan, Gould and Heckel2001; Morin et al., Reference Morin, Biggs, Sisterson, Shriver, Ellers-Kirk, Higginson, Holley, Gahan, Heckel, Carrière, Dennehy, Brown and Tabashnik2003; Xu et al., Reference Xu, Yu and Wu2005). Recently, through a biphasic F1 screening strategy, two resistance alleles of cadherin of H. armigera were identified from a field population collected from the Yellow River cotton area of China in 2005 (Yang et al., Reference Yang, Chen, Wu, Yang and Wu2007).
A disrupted allele (r 1) of a cadherin gene (Ha_BtR) is genetically associated with resistance to Cry1Ac in a Cry1Ac-selected strain GYBT of H. armigera (Xu et al., Reference Xu, Yu and Wu2005). In the present study, the r 1 allele from the GYBT strain was introgressed into a susceptible SCD strain by crossing between GYBT and SCD strains, followed by repeated backcrossing to the SCD strain and molecular marker assisted family selection. The cross-resistance pattern and inheritance mode of Cry1Ac resistance in the introgressed strain (SCD-r1) were characterized.
The SCD and SCD-r1 strains are near-isogenic lines, which are appropriate materials for studies on fitness costs associated with the Ha_BtRr 1 allele of H. armigera. Life history traits on the artificial diet and susceptibility to chemical insecticides of the susceptible SCD and resistant SCD-r1 strains were compared to determine whether there is a fitness cost associated with Cry1Ac resistance in H. armigera.
Materials and methods
Insect strains
The susceptible strain (SCD) originated from the Cote D'Ivoire in the 1970s and was passed to our laboratory by Bayer Crop Science in 2001. The SCD strain was maintained in the laboratory without exposure to insecticides or Bt toxins. The resistant GYBT strain was derived from a field-collected population (GY) from Hebei Province of China in 2001 by 28 generations of selection with activated Cry1Ac (Xu et al., Reference Xu, Yu and Wu2005). The GYBT strain has more than 500-fold resistance to activated Cry1Ac compared with the parental GY strain. The GY strain is homozygous for the wild type allele of Ha_BtR (Helicoverpa armigera cadherin-like gene), and the GYBT strain is homozygous for a truncated allele (r 1) of Ha_BtR (Xu et al., Reference Xu, Yu and Wu2005; Yang et al., Reference Yang, Chen, Wu, Yang, Xu and Wu2006).
Larvae of H. armigera were reared on an artificial diet based on wheat germ and soybean powder at 27±1°C with a 16:8 (L:D) light regime. Adults were held under the same temperature and light conditions at an RH of 60% and supplied with a 10% sugar solution.
Molecular detection of the r1 allele of Ha_BtR
Individual adults (half of the abdomen tissue) were ground in 300 μl of hot DNA lysis buffer (100 mM Tris, 50 mM EDTA, 200 mM NaCl, 1% SDS, pH 8.0). After adding 2 μl Proteinase K (20 mg ml−1), the homogenates were incubated at 56°C for 1–3 h. The homogenates were extracted twice with a solution of 150 μl phenol plus 150 μl chloroform-isoamylalchohol (24:1, v/v). After centrifugation at 12,000 rpm for 5 min, the supernatant was collected and DNA was precipitated using 900 μl 100% ethanol. The DNA pellet was recovered by centrifugation at 12,000 rpm, washed with 600 μl 70% ethanol and resuspended in 50-100 μl of TE buffer (1 mM EDTA, 10 mM Tris, pH 8.0) and stored at −20°C for use.
Allele specific PCR (AS-PCR) was used to amplify and discriminate between the wild type allele and the mutant allele (r 1) of Ha_BtR using the method of Yang et al. (Reference Yang, Chen, Wu, Yang, Xu and Wu2006). Briefly, two reverse primers were designed to cover the variation site: S-R (5′-CTATGTAGAACGCCTCGTGAG-3′, located in Exon 9) for the wild allele, and r1-R (5′-CTTCACACATGATGTTCCTCG-3′, located in Exon 25) for the mutant r 1 allele. A third primer, r1-F (5′-AGACAGGGACACTCTTGAGAAG-3′, located in Exon 8), was designed as the common forward primer. The amplification reactive mixture (25 μl) contained 100 ng genomic DNA, 1 μM of each primer, 150 μM of dNTP, 2 mM of MgCl2, 1 U of rTaq DNA polymerase and 2.5 μl of 10× PCR buffer. PCR was performed for 30 cycles of 30 s at 94°C, 1 min at 55°C, and 2 min at 72°C. PCR products were analyzed by 1.5% agarose gel electrophoresis and ethidium bromide staining.When the primer pair r1-F/r1-R was used for PCR amplification, the resistant homozygote (r 1r 1) and heterozygote (r 1S) had a 410 bp fragment, while the susceptible homozygote (SS) did not have this fragment. However, when the primer pair r1-F/S-R was used, the susceptible homozygote (SS) and heterozygote (r 1s) had a 650 bp fragment, while the resistant homozygote (r 1r 1) did not have this fragment.
Introgression of the r1 allele from the GYBT strain into the SCD strain
The program for backcrossing and family selection was illustrated in fig. 1. Males from the GYBT strain (r 1r 1) were crossed (mass mating, 50 pairs) with females from the SCD strain (SS) to produce F1 progeny (r 1S). The F1 males were crossed (mass mating, 50 pairs) with the SCD females to produce BC1 progeny (r 1S+SS, 1:1). Then, the BC1 males were backcrossed individually with the SCD females (single-pair mating). After AS-PCR detection of genotypes of the Ha_BtR gene of all BC1 males, only single-pair families derived from the r 1S father were kept to produce BC2 (r 1S+SS, 1:1). In the same way, single-pair families between the BC2 males with the r 1S allele and the SCD females were kept to produce BC3 (r 1S+SS, 1:1), and single-pair families between the BC3 males with the r 1S allele and the SCD females were kept to produce BC4 (r 1S+SS, 1:1). The males and females of BC4 were crossed to produce BC4F1 progeny (r 1r 1+r 1S+SS, 1:2:1). The BC4F1 progeny were selected with 5 μg cm−2 Cry1Ac, which is expected to kill all r 1S and SS individuals; and only r 1r 1 homozygotes were kept to generate a homozygous strain of r 1r 1 (SCD-r1). In cases where there was any ambiguity, single-pair families between SCD-r1 males and females were established, and all parents of these single-pair families were subjected to an AS-PCR examination to confirm their homogeneity for the r 1 allele of Ha_BtR gene.
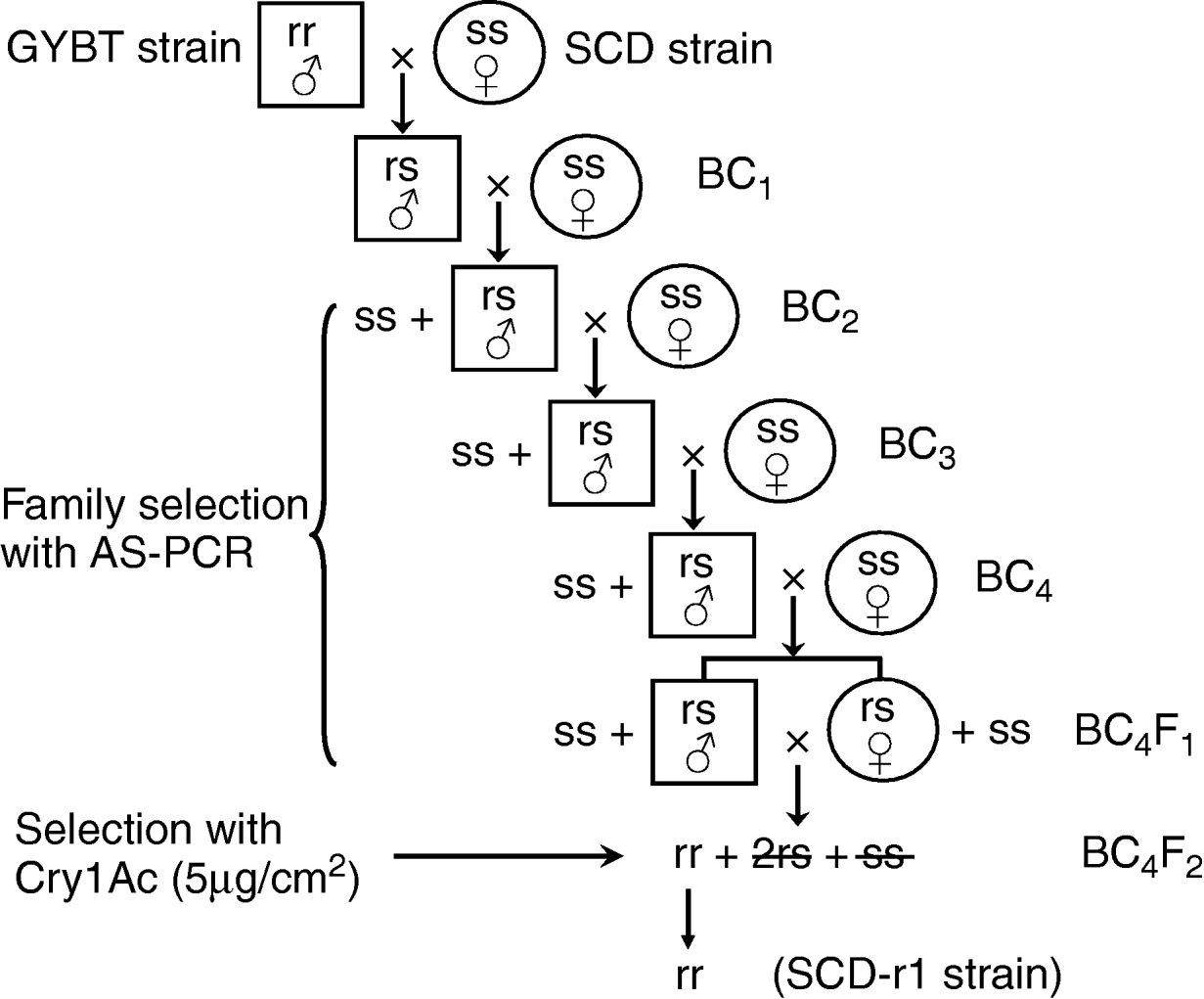
Fig. 1. Schematic program for introgression of a disrupted cadherin gene from the GKBT strain into the SCD strain of H. armigera.
Bioassay of Bt toxins and chemical insecticides
The toxicity of the various B. thuringiensis proteins to H. armigera was determined by a surface contamination bioassay. The toxin suspensions of the proteins were diluted with 0.01 M pH7.4 phosphate buffer solution (PBS) to generate five to seven serial dilutions. PBS was used as a control. A disc of artificial diet (16 mm dia.) was put into a 24-well plate and made to fit into the inner wall and bottom of the plate by gentle pressure. 100 μl of the toxin solution was applied to the diet surface and allowed to air dry. One second-instar larva, starved for four hours, was placed in each well of the plate and covered with two layers of nylon net to prevent escape. Forty-eight larvae were tested for each concentration. The mortality and body mass of the survivors were measured after five days being kept at 26±1°C with a 16:8 (L:D) photoperiod and 60% RH. Both dead larvae and those with a body mass of less than 5 mg were recorded as dead.
Technical grade fenvalerate (97%, Sumitomo, Japan) or monocrotophos (94%, Nantong Pesticide Factory, China) was dissolved in acetone and a series of concentrations was prepared. For each concentration, at least 40 third instar larvae (9–12 mg) were treated topically on the thorax with 0.25 μl insecticide by Hamilton syringe. After 48 h, the mortality was assessed. Larvae were classified as dead if they were unable to move after being prodded with a needle.
Inheritance analysis
Virgin female moths from the SCD-r1 strain were mass-crossed with male moths of the SCD strain and vice visa. The responses of F1 hybrids from the two reciprocal crosses to activated Cry1Ac were determined. The dominance of resistance was calculated using the method of Stone (Reference Stone1968). The degree of dominance (D) values range from −1 (completely recessive) to +1 (completely dominant).
Survival and development in the laboratory
For each strain, 300 newly-hatched neonates were placed individually in glass tubes (20 ml volume) with artificial diet under 26±1°C. Larval development status was individually recorded. Within 48–72 h of pupation, pupae were weighed. Emergence of adults was recorded daily and adults were paired in the oviposition cages. The eggs were collected at an interval of 24 h. The net replacement rate (R 0), the number of daughters that replace an average female over a course of a generation (Birch, Reference Birch1948), was calculated for each strain by the formula R 0=(n⋅I e·I a)/2, where n is the mean number of eggs per female, I e is the fraction of eggs which are fertile, I a is the fraction of adults which are reproductive and 2 is the sex ratio coefficient. The relative fitness was calculated as the R 0 of the resistant SCD-r1 strain divided by the R 0 of the susceptible SCD strain.
Data analysis
Bioassay data were analysed using Poloplus® software (LeOra Software, California, 2003). The mean larval development time and the pupal duration of the different strains were compared using analysis of variance (ANOVA).
Results
Introgression of the r1 allele
The r 1 allele of Ha_BtR gene was introgressed into a susceptible SCD strain by crossing between GYBT and SCD strains, followed by four iterations of backcrossing to the SCD strain. Single-pair families resulting from backcrossing were subjected to family selection with allele specific PCR, and only progeny derived from male moths heterozygous for the r 1 allele were selected for further backcrossing. Molecular detection data is detailed in table 1. Progeny (BC4F2) from single-pair families from heterozygous male and heterozygous female moths of BC4F1 were subjected to selection with 5 μg cm−2 of Cry1Ac, which is expected to kill all r 1S and SS according to previous experience from the GYBT strain (Yang et al., Reference Yang, Chen, Wu, Yang and Wu2007). Thirty-eight single-pair families produced from survivors of BC4F2 under Cry1Ac selection were established, and all parents of these single-pair families were confirmed to be homozygous for the r 1 allele of Ha_BtR gene. The strain derived from the 38 single-pair families was designated as SCD-r1. The SCD and SCD-r1 strains are a pair of near isogenic lines (NILs) for the Ha_BtR locus. It was estimated, using the principle of Mendelian segregation, that the SCD-r1 strain shared 96.7% (31/32) of its genome with that of the recipient SCD strain.
Table 1. Molecular detection of the r 1 allele of Ha_BtR gene of H. armigera.

N, Number of moths checked.
Cross resistance spectrum
After the mutated r 1 allele of Ha_BtR was introgressed into the recurrent parent SCD strain, the toxicity of Cry1Ac, Cry1Aa, Cry1Ab and Cry2Aa against the SCD-r1 and SCD strains was compared (table 2). Compared with the SCD strain, the SCD-r1 strain exhibited high levels of resistance to Cry1Ac (438-fold), medium levels of resistance to Cry1Aa (>41-fold) and Cry1Ab (31-fold), but there was no resistance to Cry2Aa (1.2-fold). This indicates that the loss of function mutation of Ha_BtR alone is enough to confer high levels of resistance to Cry1Ac and significant cross resistance to the other two Cry1A toxins tested.
Table 2. Toxicity of activated Cry1Ab, Cry1Ac and Cry2Aa against the SCD and SCD-r1 strains of H. armigera.

* LC50s for which the 95% confidence limits (95%CL) did not overlap were considered to be significantly different.
N, Number of second-instar larvae tested.
Toxicity of two chemical insecticides fenvalerate and monocrotophos against the SCD-r1 strain was tested by topical application bioassay, and the LD50 was not significantly different between the two strains (table 3), suggesting that loss of function mutation of Ha_BtR does not confer any pleiotropic effects on susceptibility of H. armigera to the two nerve-toxic insecticides.
Table 3. Toxicity of fenvalerate and monocrotophos against the SCD and SCD-r1 strains of H. armigera.

* LD50s for which the 95% confidence limits (95%CL) did not overlap were considered to be significantly different.
N, Number of third-instar larvae tested.
** Toxicity ratio=LD50 (SCD-r1)/LD50(SCD).
Inheritance mode
Toxicological responses of the resistant SCD-r1, susceptible SCD and their F1 progeny to Cry1Ac toxin are summarized in table 4. The responses to Cry1Ac of F1 progeny from reciprocal crosses between the SCD-r1 and SCD strains were not statistically different. This indicates that the Cry1Ac resistance is autosomal. Using the method of Stone (Reference Stone1968), the degree of dominance (D) in resistance was calculated to be −0.98 and −1 at LC50, suggesting that Cry1Ac resistance in the SCD-r1 strain was completely recessive.
Table 4. Toxicity of Cry1Ac against the susceptible SCD strain, resistant SCD-r1 strain and their F1 progeny of H. armigera.

N, Number of second-instar larvae tested.
Relative fitness of the SCD and SCD-r1 strains
Life tables of the SCD and SCD-r1 strains fed on artificial diet were constructed to determine net replacement rate (R 0), and no difference was observed between R 0 of the two strains (table 5).
Table 5. Life table and relative fitness of the SCD and SCD-r1strains of H. armigera reared on artificial diet.

* Relative fitness=R0 of SCD-r1 strain)/R0 of SCD strain.
The larval development time of the resistant SCD-r1 strain was significant longer than that of the SCD strain, but pupal development time of the two strains was not significantly different (table 6).
Table 6. Developmental time (day) of larvae and pupae of the SCD and SCD-r1 strains of H. armigera.

N, Number of evaluated individuals for each strain.
* Means between strains followed by the different letters are significantly different (P=0.05).
Discussion
Repeated backcrossing has been frequently applied to the genetic analysis of insecticide resistance. Not only can this method be useful in the genetic analysis of resistance, it can be used to move a major resistance gene into a susceptible genetic background and, thereby, isolate it from other genes that affect the resistance phenotype (Roush & Daly, Reference Roush, Daly, Roush and Tabashnik1990). Marker-assisted backcrossing is routinely applied for gene introgression in plant and animal breeding (Frisch & Melchinger, Reference Frisch and Melchinger2005), but it is infrequently used in the study of insecticide resistance. In the current study, a marker-assisted backcrossing regime worked successfully to introduce a deletion mutation allele of Ha_BtR into a susceptible SCD strain. The results further confirmed that the loss of function mutation of the Ha_BtR gene alone can confer resistance to Cry1Ac and cross resistance to Cry1Aa and Cry1Ab. Cry1Ac resistance in the GYBT strain of H. armigera is incompletely recessive, but it is completely recessive in the SCD-r1 strain. Levels of cross resistance to Cry1Ab and Cry1Aa of the SCD-r1 strain differed a little to those of the GYBT strain. These indicated some minor factor or factors affecting resistance phenotype were removed through the repeated backcross regime employed in the present study.
Since the first Cry toxin receptors were cloned in 1995, much progress has been made in our understanding of toxin specificity and mode of action (Pigott & Ellar, Reference Pigott and Ellar2007). The best-characterized receptors of Cry1A toxins are aminopeptidase N (APN) proteins and cadherin proteins from lepidopterans. Bravo et al. (Reference Bravo, Gill and Soberón2007) proposed that both cadherin and APN receptors are required for full Cry1A toxicity against Manduca sexta. The toxin and receptor binding is suggested to be sequential; an activated toxin monomer binds initially to a cadherin receptor (BT-R1) and then to a second receptor (class 1 APN). The cadherin binding results in a conformational change of activated Cry1Ac that facilitates cleavage of helix α−1 from Cry1Ac domain 1 by a membrane-bound protease. Further activated toxin oligomerizes to form a tetrameric prepore and subsequently binds to APN, which finally results in pore formation and intoxication. This model is consistent with the finding of Chen et al. (Reference Chen, Hua, Jurat-Fuentes, Abdullah and Adang2007) that peptide fragments from membrane-proximal regions of the cadherin protein (Bt-R1) unexpectedly increase the toxicity of Cry1A. These fragments could accelerate the hypothesized additional toxin processing step. A recent study reported that modified Bt Cry1A toxins (Cry1AbMod and Cry1AcMod) lacking one α-helix did not require cadherin to form oligomers, and the modified Cry1A toxins killed cadherin-silenced M. sexta and Bt-resistant P. gossypiella that had cadherin deletion mutations (Soberón et al., Reference Soberón, Pardo-López, López, Gómez, Tabashnik and Bravo2007). It would be interesting to test the concept of the mode of action of modified Cry1A in the SCD-r1 and SCD strains of H. armigera developed in the present study to determine how universal the effects of mutations in cadherins are.
Pleiotropy is the ability of a single allele to have more than one distinguishable effect. Selection for insect resistance to insecticides generally results in a reduction in the fitness of the resistant insects, which is caused by the antagonistic pleiotropy of the resistant alleles (Roush & Daly, Reference Roush, Daly, Roush and Tabashnik1990; Chevillon et al., Reference Chevillon, Denis, Rousset, Pasteur and Raymond1997; Gould, Reference Gould1998). In the absence of the pesticide selection, resistant strains of insects often show fitness costs, such as longer developmental time (Oppert et al., Reference Oppert, Hammel, Thorne and Kramer2000; Carrière et al., Reference Carrière, Ellers-Kirk, Biggs, Nyboer, Unnithan, Dennehy and Tabashnik2006), lower fertility and fecundity (Alyokhin & Ferro, Reference Alyokhin and Ferro1999), lower emergence from diapause (Carrière et al., Reference Carrière, Ellers-Kirk, Patin, Sims, Meyer, Liu, Dennehy and Tabashnik2001) and decreased ability to response to alarm pheromones (Foster et al., Reference Foster, Denholm, Thompson, Poppy and Powell2005) or to avoid predation (Agnew et al., Reference Agnew, Berticat, Bedhomme, Sidobre and Michalakis2004; Berticat et al., Reference Berticat, Duron, Heyse and Raymond2004).
However, it is often difficult to associate fitness disadvantages specifically with resistance, for resistant and susceptible strains may differ in fitness attributes independently of resistance (Roush & Daly, Reference Roush, Daly, Roush and Tabashnik1990). The extent of fitness costs could have been overestimated or underestimated in studies based on comparisons between strains, and overestimation would have been more common if selection for resistance reduces population size and accelerates accumulation of deleterious mutations in resistant strains (Carrière et al., Reference Carrière, Ellers-Kirk, Biggs, Nyboer, Unnithan, Dennehy and Tabashnik2006). So, it is critical to compare all three genotypes (rr, rs, ss) in a similar genetic background, preferably in a strain that has been repeatedly backcrossed to a strain without previous pesticide exposure. Bird & Akhurst (Reference Bird and Akhurst2004, Reference Bird and Akhurst2005) minimized divergence between strains by using four generations of serial backcrossing to increase isogenicity of their Bt-susceptible and Bt-resistant strains of H. armigera. Carrière et al. (Reference Carrière, Ellers-Kirk, Biggs, Nyboer, Unnithan, Dennehy and Tabashnik2006) used hybrid strains of P. gossypiella to generate a similar genetic background for Bt resistance genotypes. In the current study, a pair of near-isogenic strains of the SCD-r1 and SCD strains of H. armigera was developed to homogenize the genetic background other than the resistance locus. The fitness cost of slower larval development in the resistant strain appears, therefore, to be associated with a specific resistance mechanism (loss of function mutation of Ha_BtR).
Fitness costs imposed by resistance genes can be magnified under stressful conditions, such as poor quality food resources or harsh weather conditions. The fitness costs associated with Bt resistance in Trichoplusia ni, H. armigera and P. xylostella increased with declining host plant suitability (Janmaat & Myers, Reference Janmaat and Myers2005; Bird & Akhurst, Reference Bird and Akhurst2007; Raymond et al., Reference Raymond, Sayyed and Wright2007). Fitness costs of resistance to spinosad in P. xylostella increased in scale at unfavourably low and high temperatures (Liu et al., Reference Liu, Liu, Liu and Ye2007). The current study investigated the fitness costs of Bt-resistant H. armigera under optimized conditions (reared on artificial diet in the laboratory); further work on relative fitness of the SCD and SCD-r1 strains of H. armigera under field conditions is ongoing.
Acknowledgements
This research was supported by grants from the National Basic Research Program of China (2007CB109204), National Natural Science Foundation of China (30571233), National High Technology Research and Development Program of China (2007AA10Z420) and the 111 project (B07030).









