Introduction
Filarioids (Filarioidea) are heteroxenous parasitic nematodes of tissues and body cavities of many vertebrate hosts, except fish. Members of the family Onchocercidae produce blood- or skin-inhabiting microfilariae that are ingested by blood-feeding arthropods and undergo a two-stage development until the third stage (L3) is transmitted to a new receptive host. More than 30 species of filarioids have been found in wild primates and humans in the Neotropical forests (Bain et al., Reference Bain, Baker and Chabaud1982). Of these, 13 species belong to the genus Mansonella (Tetrapetalonema) (Faust, 1935) and six to the genus Dipetalonema (Diesing, 1861), which are restricted to platyrrhine (Neotropical) primates (Bain et al., Reference Bain, Mutafchiev, Junker, Guerrero, Martin, Lefoulon and Uni2015; Laidoudi et al., Reference Laidoudi, Medkour, Levasseur, Davoust and Mediannikov2020). Biting midges (Diptera: Ceratopogonidae) are competent vectors of Dipetalonema spp. as demonstrated for Culicoides hollens for Dipetalonema caudispina (Molin, 1858) and Dipetalonema gracile (Rudolphi, 1809) (Eberhard et al., Reference Eberhard, Lowrie and Orihel1979).
Overall, the genus Dipetalonema includes: D. caudispina (Molin, 1858), Dipetalonema freitasi Bain, Diagne and Muller, Reference Bain, Diagne and Muller1987, D. gracile (Rudolphi, 1809), Dipetalonema graciliformis Freitas, 1964, Dipetalonema yatesi Notarnicola, Jimenez and Gardner, Reference Notarnicola, Jiménez and Gardner2007 and Dipetalonema robini Petit, Bain and Roussilhon, Reference Petit, Bain and Roussilhon1985 (Table S1). Adhering to the concept of morphological species, the six species above are mainly distinguished by the complex structures of the vagina vera, caudal lappets in females as well as the spicules, the area rugosa of the tail and arrangement of the musculature in males and the shape of microfilariae (Petit et al., Reference Petit, Bain and Roussilhon1985; Bain et al., Reference Bain, Diagne and Muller1987).
Dipetalonema spp. have been reported from at least 20 species of Neotropical primates, from southern Mexico to central Bolivia and subtropical Brazil and Argentina (see Table S1). The holotype of D. graciliformis from Saguinus midas in Brazil (Pará State) was deposited by Freitas and described morphologically by Bain et al. Reference Bain, Petit and Rosales-Loesener1986, along with specimens collected in French Guiana. Similarly, two species of Dipetalonema (i.e. D. gracile and D. robini) were described in Saimiri sciureus from Guyana (Petit et al., Reference Petit, Bain and Roussilhon1985) indicating the sympatric distribution of Dipetalonema spp. in this geographic region. Later on, D. graciliformis was found in the abdominal cavity of Saguinus mystax (Spix, 1823) from Amazonas State, Brazil and of Saguinus labiatus in Peru whereas D. freitasi from Cebus capucinus in captivity and D. yatesi in Ateles chamek from north-central Bolivia.
Here, we performed integrative studies combining morphological and molecular data on filarioid nematodes isolated from the peritoneal cavity of red-handed tamarins (S. midas, Linnaeus 1758).
Materials and methods
Adult filarioid worms (nine females and five males) were retrieved from the peritoneal cavities (Fig. S1) of two of the four red-handed tamarins found road killed on the edge of the national road 1, near the coast of northern French Guiana. Collected worms were identified at the species level based on morphological criteria. In addition, three molecular loci (i.e. cox1, 18S and 28S genes) were amplified, sequenced and deposited in the GenBank database under the accession numbers: MW357380-357394, MW355926-355940 and MW355911-355925 for the cox1, 18S and 28S genes, respectively (see Supplementary File S1 for details).
Results
Species identification and general morphological description
All filarioids were identified as D. graciliformis, according to Petit et al. (Reference Petit, Bain and Roussilhon1985) and Bain et al. (Reference Bain, Petit and Rosales-Loesener1986). The comparative measurements of adult females and males from the current study and the other Dipetalonema species are detailed in Tables 1 and 2. Body whitish with fine transverse cuticular striation (Fig. S2A). Anterior end rounded (Fig. S2B). Buccal capsule small with head cuticle smooth cephalic region with smooth quadrangular shape (Fig. S2B). Oral opening small with four labial papillae arranged on a laterally elongated rectangle and four cephalic papillae located around oral opening. The oesophagus is divided in an anterior muscular part (short) (Fig. S2C) and posterior glandular portion (long) (Fig. S2C). The posterior end tapering to a point (Fig. S2D). Nerve ring located at half-length of muscular oesophagus, the excretory pore is visible behind the nerve ring. Vulva at the level of the oesophagus. Ovaries are located at the mid-body (Fig. 1A). Microfilaria (Fig. 1B) were obtained from the uteri (Fig. 1C) of a mature female. The female presented with extremity with three well developed petaloid appendage triangular shape with side base (Fig. 1D).
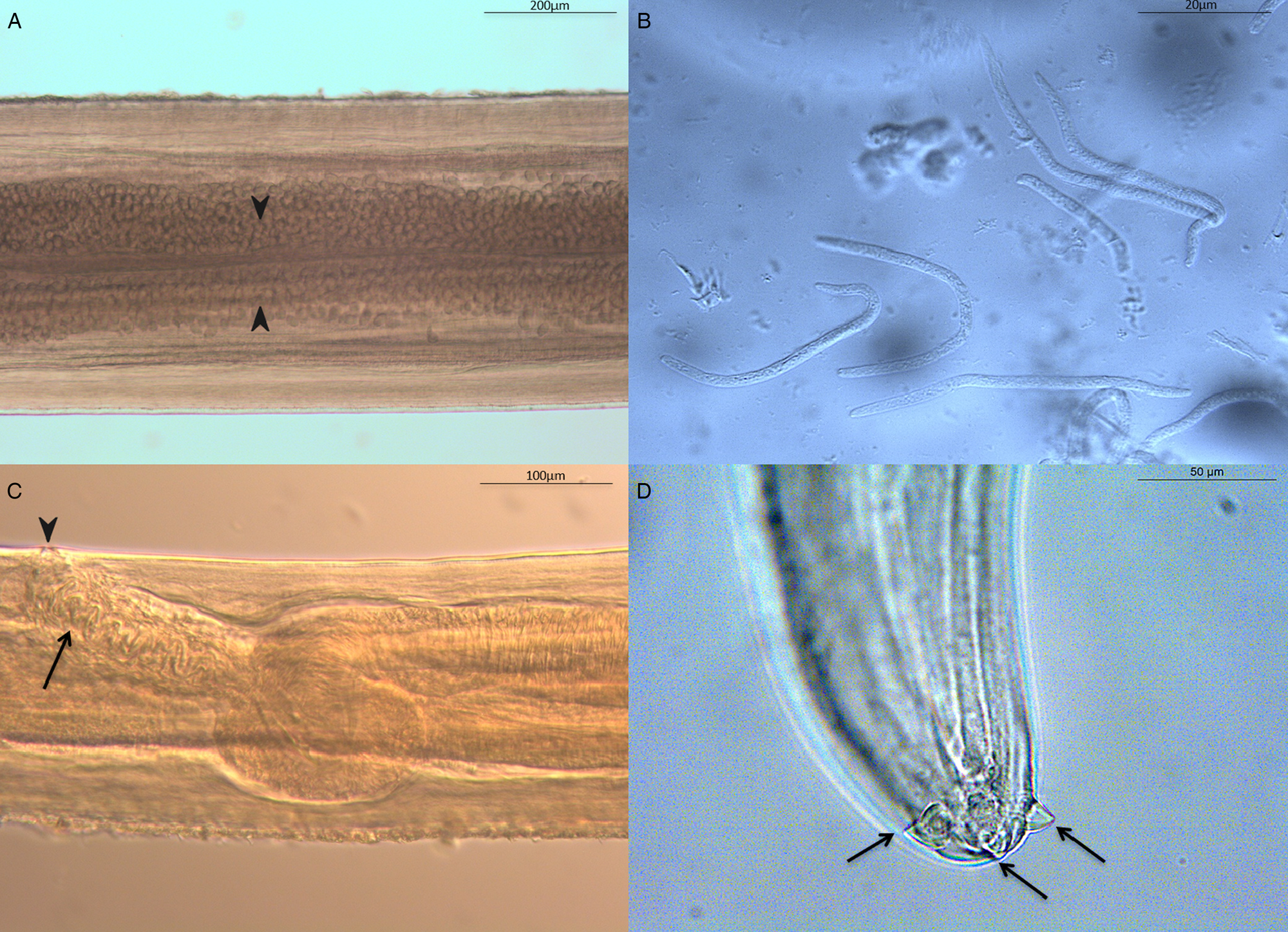
Fig. 1. Morphology of Dipetalonema graciliformis female. (A) Anterior part of female body showing ovary structure to the mid-body (arrows). (B) Uterine microfilaria. (C) Lateral view of the anterior region showing: genital opening (arrowhead), vagina vera (anterior end) with sinuosis duct (arrow). (D) Posterior end (ventral view) with details of the lappets.
Table 1. Comparative measurement (in μm unless specified) of adult females of Dipetalonema graciliformis (Freitas, 1964) from our study with the six Dipetalonema species

Table 2. Comparative measurement (in μm unless specified) of adult males of D. graciliformis (Freitas, 1964) from our study with the six Dipetalonema species

Caudal extremity in male spirally coiled accompanied by caudal papillae (Fig. S3A). The male presents single testis at the level of the oesophago-intestinal junction. The paired spicules are unequal in size being the left longer than the right one (Fig. S3A); the left spicule subdivided into the proximal handle (Fig. S3B) and lamina (Fig. S3C), being the latter formed by a membranous alae and a flagellum (Fig. S3D). Laterally (Fig. S4A), it is possible to observe: three pair of the pre-cloacal papillae, a single central pre-cloacal papilla, a pair of ad-cloacal papillae and one pair of post-cloacal papillae (the second pair was not visible) plus four papillae at the end of the tail near to the caudal appendages shaped like a nipple (Fig. S4B). A very small gubernaculum (Fig. S4C) is of navicular shape with post cloacal bands present. Posterior extremity with two spiral twist with small longitudinal striae as transverse band on ventral surface representing the area rugosa (Fig. S4D) formed by rows of short longitudinal crest. Body size ratio of female to male 1:1.8.
Molecular phylogenetic analyses
All polymerase chain reaction amplifications yielded the DNA amplicon of expected size from all the 14 specimens examined. Sequence alignment showed the identity of all DNA amplicons for each gene. No gaps were found in the alignment of the cox1 gene against the reference mitogenome of Acanthocheilonema viteae (HQ186249), and when translated, there were no stop codons in the amino acid sequences, suggesting the absence of co-amplified numts. BLASTn analysis revealed that the nucleotide and aminoacid sequences were 98.65 and 100% homologous with those from D. graciliformis (GenBank accession number: KP760182, Protein Id. ALR73841, voucher 220YU ind1 MNHN) suggesting that nucleotide mutations are silent. In addition, the BLASTn analysis of the 18S rDNA sequence revealed 100% of identity with the GenBank sequence (GenBank accession number: MT336175) of a genotype isolated from howler monkey in the same area, previously referred to as an unidentified Onchocercidae species (Laidoudi et al., Reference Laidoudi, Medkour, Levasseur, Davoust and Mediannikov2020).
The maximum liklihood trees inferred from cox1 (Fig. 2A), COX1 (Fig. 2B) and the concatenated sequences (cox1 and 18S) (Fig. S5) produced a similar topology and very similar posterior bootstrap values. The trees provide evidence that investigated filarioids are an integral part of the Onchocercidae clade 4 (ONC4) (Fig. 2A and 2B and Fig. S5). This organism clustered with D. graciliformis (GenBank accession number: cox1: KP760182, COX1: Id. ALR73841, 18S: KP760131, voucher 220YU ind1 MNHN) in all phylograms. Similarly, the lowest interspecific nucleotide pairwise distance was observed with D. graciliformis (Fig. S5). BLAST search revealed that Onchocercidae members, in particular those of clade 4 (ONC4) lacked the representative 28S in both GenBank and Worm–Parasite databases. The 28S ML tree allowed the comparison of D. graciliformis with members representing three Onchocercidae (ONC2, ONC3 and ONC5) clades. The phylogram supported the divergence of D. graciliformis from these clades (Fig. S6).

Fig. 2. Comparative phylograms showing the position of D. graciliformis (indicated in red) among Onchocercidae Clades. ML inferences were generated from (A) the partial (431-bps) nucleotide sequences of the cox1 gene and (B) 159-aa from their translated (COX) protein sequences. The axis shows the global distance observed throughout the trees. Branches are colour-coded according to the bootstrap's values. The identity of each taxa is colour-coded according to the Onchocercidae Clades. Outgroup taxons are showing in grey. GenBank accession numbers, protein Id, species name and hosts are indicated at each node. The number of base substitutions per site between D. graciliformis isolated in the current study and the other Onchocercidae members are shown for both cox1 and COX1 sequences. Log-likelihood were −6216.4 and −1793.4 for the cox1 and COX1 sequences, respectively.
Discussion
Neotropical primates are commonly parasitized by filarioid nematodes of the genera Dipetalonema and Mansonella (Bain et al., Reference Bain, Mutafchiev, Junker, Guerrero, Martin, Lefoulon and Uni2015; Laidoudi et al., Reference Laidoudi, Medkour, Levasseur, Davoust and Mediannikov2020). Many of these primates are threatened due to anthropic pressures such as the illegal wildlife trade (Bezerra-Santos et al., Reference Bezerra-Santos, Mendoza-Roldan, Thompson, Dantas-Torres and Otranto2021). These parasites are living in various tissues and cavities outside the gastrointestinal tract (Strait et al., Reference Strait, Else and Eberhard2012), and they may induce pathologies that involve pleuritis, fibrinopurulent peritonitis and fibrinous adhesion, resulting in the entrapment of worms (Baker, Reference Baker, Robert, Lynn, Suzette, Keith and James2018). Thus, understanding the impact of parasitic helminths on their primate hosts is an aspect that should not be overlooked as a vital part of primate conservation. Studies on parasitic nematodes of primates are scant in the Neotropics as it is difficult to obtain parasites from these highly mobile, arboreal and protected animals. The filarioids reported herein were recovered from the abdominal cavity of tamarins found road-killed, which documents the high value of such a material for studies in pathogens of wildlife. The current study emphasizes the usefulness of the combined morphological and molecular-based approaches in identifying filarioid parasites. Morphological and molecular data prove the conspecificity of the retrieved material with D. graciliformis, confirming the presence of this species in free-ranging red-handed tamarins (Bain et al., Reference Bain, Petit and Rosales-Loesener1986) and howler monkeys (Laidoudi et al., Reference Laidoudi, Medkour, Levasseur, Davoust and Mediannikov2020).
The genus Dipetalonema is restricted to Neotropical primates (Lefoulon et al., Reference Lefoulon, Bain, Bourret, Junker, Guerrero, Cañizales, Kuzmin, Satoto, Cardenas-Callirgos, de Souza Lima, Raccurt, Mutafchiev, Gavotte and Martin2015) with a high species diversity observed in a wide range of host species (Table S1). Morphologically, D. graciliformis is close to D. gracile, though females of D. graciliformis are longer, present shorter oesophagus and tail and slightly shorter microfilariae with two obtuse extremities (Bain et al., Reference Bain, Petit and Rosales-Loesener1986). The male of D. graciliformis can be distinguished from D. gracile by its caudal region which is coiled on three turns of spires and a longer flagellus in the longer spiculum (Bain et al., Reference Bain, Petit and Rosales-Loesener1986).
The cox1 analysis allowed us to discriminate D. graciliformis. We used three different methods for the cox1 analysis, which confirmed that the filarioids studied clustered with D. graciliformis isolated from Saimiri sciureus in Peru (GenBank accession number: KP760182). The cox1 gene is an efficient DNA barcode for filarioid species which is due to its low nucleotide distances (from 0 to 0.02) within filarioid species (Ferri et al., Reference Ferri, Barbuto, Bain, Galimberti, Uni, Guerrero, Ferté, Bandi, Martin and Casiraghi2009) and a larger variation between congeneric species (i.e. 0.098–0.2) (Casiraghi et al., Reference Casiraghi, Anderson, Bandi, Bazzocchi and Genchi2001; Ferri et al., Reference Ferri, Barbuto, Bain, Galimberti, Uni, Guerrero, Ferté, Bandi, Martin and Casiraghi2009). In addition, the cox1 has been successfully used to monitor the distribution of emerging nematodes, such as the case of Thelazia callipaeda eyeworm (Otranto et al., Reference Otranto, Mendoza-Roldan and Dantas-Torres2021). On the other hand, the 18S rRNA gene has been proven a proper target to reconstruct the phylogenetic history of nematode clades (Blaxter et al., Reference Blaxter, De Ley, Garey, Llu, Scheldeman, Vierstraete, Vanfleteren, Mackey, Dorrls, Frisse, Vida and Thomas1998). Despite the absence of representative 18S rDNA data with high query cover, phylogenetic analysis from the concatenated partial 18S and cox1 sequences clustered the filarioids from the current study with D. graciliformis (GenBank accession number: KP760182 and KP760131 for the cox1 and the 18S, respectively). In contrast, the analysis of the 28S was not sufficiently conclusive, which is due to the lack of representative data from GenBank and Worm databases.
In the current study, we provide detailed morphological data on D. graciliformis isolated from red-handed tamarins from French Guiana, as well as analyses of nuclear and mitochondrial molecular markers. The analysis of 18S sequences from examined nematodes proven their identity with previously reported undescribed onchocercid filarioids from howler monkeys from the same area (Laidoudi et al., Reference Laidoudi, Medkour, Levasseur, Davoust and Mediannikov2020). Further studies are needed to clarify the epidemiology, circulation and host diversity of these parasites as well as their possible impact on the fitness of infected individuals of Neotropical primates.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182021000901
Author contributions
YL, OM, DO and BD conceived and designed the study. BD and CAB conducted the fieldwork. YL and RPL performed laboratory analysis. YL, JAMR, DM and DO wrote the article.
Financial support
This study was supported by the French Military Health Service and the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the programme ‘Investissements d'avenir’, reference ANR-10-IAHU-03, the Région Provence-Alpes-Côte d'Azur and European funding FEDER PRIMI.
Conflict of interest
None.