Introduction
Analysis of population genetic structure may provide insight into aspects such as the scale at which populations are structured, the development of host strains and the impact of environmental factors. These aspects are essential for understanding the ecology of insect agricultural pests since they are often implicit in the management of population levels. A comparative approach may be useful to determine the factors most influential in structuring populations of a taxon. Here, we use such an approach to investigate South African populations of economically important Tortricidae, a cosmopolitan family containing some of the most damaging insect agricultural pest species in the world (Horak & Brown, Reference Horak, Brown, Van der Geest and Evenhuis1991).
Recently, the population genetic structure of four economically important Tortricidae in South Africa was investigated: the codling moth C. pomonella (Timm et al., Reference Timm, Geertsema and Warnich2006a), the litchi moth Cryptophlebia peltastica (Meyrick), the macadamia nut borer Thaumatotibia batrachopa (Meyrick) (Timm et al., Reference Timm, Geertsema and Warnich2006b) and the oriental fruit moth G. molesta (Timm et al., Reference Timm, Geertsema and Warnich2008). For all four species, the population genetic structure was unexpected but remarkably similar, with populations significantly structured not only at regional scale but also split into many isolated populations at local scales, such as farms or orchards. Little evidence was found to suggest that host strains of the four species had evolved in South Africa. Our aim with this study was to determine whether these patterns were also evident in the remaining two tortricid species of major economic importance in South Africa, the pear leafroller Epichoristodes acerbella (Walker) and the false codling moth T. leucotreta (Meyrick), and to use this information to further investigate which factors likely play a role in determining the population genetic structure of the Tortricidae in South Africa.
The economically important Tortricidae in South Africa are closely related and show similarities in their biology and ecology (Horak & Brown, Reference Horak, Brown, Van der Geest and Evenhuis1991; Komai, Reference Komai1999; Timm et al., Reference Timm, Geertsema and Warnich2007). Tortricid larvae are typically concealed feeders, often boring into fruit and stems and spinning and rolling leaves in which they develop. The most marked differences among the economically important species in South Africa are their population history (whether native or introduced) and host range (whether oligophagous or polyphagous). A few Tortricidae, such as C. pomonella and G. molesta, are highly successful invaders and have become established throughout the world, causing heavy economic losses wherever they are found. Other species, such as C. peltastica, T. batrachopa, T. leucotreta and E. acerbella, which are native to sub-Saharan Africa and surrounding islands, have a more local distribution, which is restricted through strict quarantine practices (Myburgh & Basson, Reference Myburgh and Basson1961; Oliver & Bolton, Reference Oliver and Bolton1974; Quilici et al., Reference Quilici, Verbizier, Trahais and Manikom1988; Newton, Reference Newton, Bedford, van den Berg and de Villiers1998). However, E. acerbella has become established in Europe as a pest of carnations (Allen, Reference Allen1980; Van der Vrie, Reference Van der Vrie, Van der Geest and Evenhuis1991). One of the factors contributing to the success of the Tortricidae is their host range. Species, such as C. pomonella, G. molesta and T. batrachopa, are limited to three or fewer hosts belonging to a single family in South Africa (Annecke & Moran, Reference Annecke and Moran1982; Blomefield, Reference Blomefield1989; De Villiers, Reference De Villiers, van den Berg, de Villiers and Joubert2001), whereas others, such as C. peltastica, E. acerbella and T. leucotreta, are extremely polyphagous (Annecke & Moran, Reference Annecke and Moran1982; Wright, Reference Wright1995; Newton, Reference Newton, Bedford, van den Berg and de Villiers1998). One of the most extreme examples of polyphagy is T. leucotreta, whose larval host range extends to at least 21 cultivated and 14 indigenous wild host plants and is, thus, one of the most economically important agricultural pests in Africa (Schwartz, Reference Schwartz1981; Newton, Reference Newton, Bedford, van den Berg and de Villiers1998).
In the few studies available on Tortricidae population genetics, which have mainly focused on C. pomonella, it is evident that the choice of marker is an important factor to consider when designing such studies. DNA-based markers, such as mitochondrial genes (Meraner et al., Reference Meraner, Brandstätter, Thaler, Aray, Unterlechner, Niederstätter, Parson, Zelger, Dalla Via and Dallinger2008) and microsatellites (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007; Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramirez2008), were unable to distinguish between populations, with one exception (Chen & Dorn, Reference Chen and Dorn2009). However, amplified fragment length polymorphism (AFLP) analysis was able to detect differences between populations, even over local scales (Timm et al., Reference Timm, Geertsema and Warnich2006a; Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008). These results agree with studies of other taxa, where AFLP analysis has been shown typically to detect high levels of genetic variation among closely related populations, particularly over small spatial scales (Jakše et al., Reference Jakše, Kindlhofer and Javornik2001; Gaudeul et al., Reference Gaudeul, Till-Bottraud, Barjon and Manel2004; Garoia et al., Reference Garoia, Guarniero, Grifoni, Marzola and Tinti2007). Thus, to facilitate studies of variation and to enable direct comparison among South African Tortricidae previously studied, we selected AFLP analysis for analyzing E. acerbella and T. leucotreta.
Material and methods
Insect material
Insect specimens were obtained by collecting infested fruit and allowing the moths to emerge or by pheromone collection. Epichoristodes acerbella specimens (n=113) were collected from seven regions in the Western Cape province, the main site of the deciduous and cut flower industries in South Africa, from commercial plantings of grapes, pears, Protea spp. and Leucadendron spp. (table 1, fig. 1). In analyses of host variation, only populations collected from grapes and pears were considered since limited numbers of individuals were collected from Protea spp. and Leucadendron spp. To assess variation over local scales, populations were collected from three sites in Stellenbosch (n=27) and from 11 different farms in the Hex River Valley (n=45; table 1). Thaumatotibia leucotreta specimens (n=163) were collected from the Western Cape, Eastern Cape and Mpumalanga provinces, which are the principal areas of tropical and subtropical fruit production in South Africa (table 2, fig. 1). From the Western Cape, individuals (n=100) were collected from six regions from citrus, pear, apple and plum orchards, as well as from acorns and pheromone traps. Populations were collected from citrus orchards in the Eastern Cape (n=19) and citrus, litchi, macadamia, star fruit orchards and pheromone traps in Mpumalanga (n=45). Due to limited population sizes collected from macadamias and star fruit, only individuals collected from citrus, pears, apples, plums, acorns and litchis were included to estimate variation among populations collected from different hosts. To obtain a measure of genetic variation over local scales, individuals were collected from a minimum of each of four different farms in Citrusdal (n=50), Stellenbosch (n=24), the Sundays River Valley (n=19) and Nelspruit (n=26; table 2). As outgroup, ten individuals obtained from Africa and maintained in the UK were used. Voucher material of specimens included for analysis was deposited in the museum of the Department of Conservation Ecology and Entomology, University of Stellenbosch.

Fig. 1. Map of South Africa showing collection details of E. acerbella and T. leucotreta populations used for AFLP analysis (•, T. leucotreta collection sites; ![]() , E. acerbella collection sites).
, E. acerbella collection sites).
Table 1. Collection details of E. acerbella specimens from South Africa used for AFLP analysis.

Brackets indicate that specimens were collected from residential areas. Hosts are listed as unknown if pheromone traps associated with multiple hosts were used for collection.
Table 2. Collection details of T. leucotreta specimens from South Africa used for AFLP analysis.

Brackets indicate that specimens were collected from residential areas. Hosts are listed as unknown if pheromone traps associated with multiple hosts were used for collection.
DNA extraction
DNA was extracted from the head and legs of moths or, in rare instances, portions of the abdomens of larvae using a CTAB-based protocol (Reineke et al., Reference Reineke, Karlovsky and Zebitz1998).
AFLP analysis
The AFLP procedure was performed as originally described (Vos et al., Reference Vos, Hogers, Bleeker, Rejans, van de Lee, Hornes, Frijters, Pot, Peleman, Kuiper and Zabeau1995), with minor modifications. Briefly, DNA was digested with the enzymes EcoRI and MseI and pre-amplification of DNA templates was performed with primers containing no selective nucleotides. Selective amplifications were performed with 33P-labelled EcoRI primers using five combinations of primer pairs, each containing three selective nucleotides. To test marker reproducibility, DNA was extracted from the same individual on different occasions and used to evaluate each selective primer pair at least three times. Selective amplification products were electrophoresed on 6% (w/v) denaturing polyacrylamide gels at 60 W for 2–3 h. Gels were dried on Whatmann paper and exposed to Kodak Biomax x-ray films for visualization.
Statistical analysis
Due to the nature of AFLP markers and protocols used for analysis, fragments were assumed to be homologous if they were of the same size. In addition, a present fragment was assumed to be dominant to an absent fragment. In this manner, AFLP profiles were recorded manually in a binary matrix using ‘1’ to denote fragment presence and ‘0’ to denote fragment absence. The so-called band-based approach (Bonin et al., Reference Bonin, Ehrich and Manel2007), rather than analyses based on allele frequency, was generally used to analyse AFLP data. The band-based approach is more suitable for analysing dominant data since it reduces preliminary assumptions used to calculate allele frequencies. In addition, since analyses are conducted at an individual level, they are more suitable for small population sizes (Bonin et al., Reference Bonin, Ehrich and Manel2007), such as those used in the current study. For all calculations, populations were assumed to be in Hardy-Weinberg equilibrium unless otherwise stated.
Genetic diversity within each population was expressed using the Shannon-Weaver information index (I) (Shannon & Weaver, Reference Shannon and Weaver1949), which does not assume that populations are in Hardy-Weinberg equilibrium but presupposes that estimates based on the numbers of bands that are present/absent are an estimate of genetic diversity (Whitkus et al., Reference Whitkus, de le Cruz, Mota-Bravo and Gomez-Pompa1998). The Shannon-Weaver index was calculated as ![]() with p i being the frequency of a given AFLP fragment (Lewontin, Reference Lewontin1972) using popgene version 1.31 software for population genetic analysis (Yeh & Yang, Reference Yeh and Yang1997).
with p i being the frequency of a given AFLP fragment (Lewontin, Reference Lewontin1972) using popgene version 1.31 software for population genetic analysis (Yeh & Yang, Reference Yeh and Yang1997).
The partitioning of genetic variability within and among populations was determined using an AMOVA analysis based on a Euclidean distance metric (Huff et al., Reference Huff, Peakall and Smouse1993), using the software GenAlEx version 6 (Peakall & Smouse, Reference Peakall and Smouse2005) based on 1000 permutations. Among populations, variability based on AMOVA analysis was used to determine the ΦPT estimator, which is analogous to F ST and can be used as a measure of population differentiation for binary data. GenAlEx version 6 (Peakall & Smouse, Reference Peakall and Smouse2005) was also used to conduct a Mantel test (Mantel, Reference Mantel1967) to evaluate correlations between the geographic origin of populations and their genetic distances, using pairwise genetic and geographic distance matrices (Smouse et al., Reference Smouse, Long and Sokal1986; Smouse & Long, Reference Smouse and Long1992). The relationships between populations were determined based on pair-wise measures of genetic distance D (Nei, Reference Nei1978; Lynch & Milligan, Reference Lynch and Milligan1994) with 1000 replications, using the software AFLP-SURV (Vekemans et al., Reference Vekemans, Beauwens, Lemaire and Roldan-Ruiz2002) with default options. Subsequently, these genetic distances were used to construct unrooted neighbour-joining trees with Phylip version 3.6 software (Felsenstein, Reference Felsenstein2004). Relationships among individuals were viewed using the unweighted pair group means arithmetic (UPGMA), based on a dissimilarity matrix generated using Jaccard's general similarity co-efficient (Sneath & Sokal, Reference Sneath and Sokal1963), with MVSP Version 3.11c (Kovach, Reference Kovach1999). When using binary data, Gower's general similarity coefficient (GGScij) is equivalent to Jaccard's co-efficient, where GGScij=a/(a+b+c), where a is the number of bands shared by individuals i, and j, b is the number of bands present in i but not in j, and c is the number in j but not in i.
Population subdivision was also tested using the program structure version 2.2 (Falush et al., Reference Falush, Stephens and Pritchard2007), which employs a model-based clustering algorithm to assign individuals probabilistically to populations (K) based on their AFLP banding profiles. AFLP data was coded as diploid individuals with missing data in one copy of all loci, according to the software manual recommendations for analysing dominant data. To estimate the number of populations, four independent runs were performed using the admixture model with an initial burn-in of 10,000 generations and 10,000 Markov Chain Monte Carlo repetitions and assuming correlated allele frequencies.
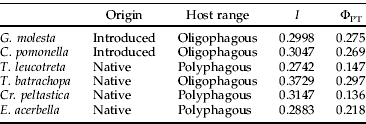
The amount and distribution of genetic variation was compared among C. pomonella, G. molesta, C. peltastica, T. batrachopa, T. leucotreta and E. acerbella using a Chi squared test. To determine whether the genetic parameters ΦPT and I differed significantly between introduced and native species as well as between oligophagous and polyphagous species, single factor ANOVA analysis was used. Species were classified as oligophagous if the larvae were able to feed on hosts belonging to a single family or as polyphagous if hosts from more than one family were known to be utilized in South Africa (table 3).
Table 3. Comparative population genetic parameters for six economically important species collected from South Africa, based on AFLP analysis.
Results
Gene flow among geographic regions
AFLP analysis using five primer combinations yielded a total of 228 fragments for analysis of E. acerbella populations and 323 fragments for analysis of T. leucotreta populations. Genetic diversity within the E. acerbella population in the Western Cape was calculated as I=0.288. The degree of genetic diversity within the T. leucotreta population was similar to that of E. acerbella, with I=0.274.
Despite the relatively high levels of genetic diversity found within total and regional populations, evidence of population differentiation in these scales was apparent in E. acerbella populations (ΦPT=0.218, P=0.001) as well as T. leucotreta populations (ΦPT=0.147, P=0.001) according to population differentiation values. For E. acerbella populations, cluster analysis (fig. 2) revealed that populations clustered broadly into four groups. The likelihood of the number of populations was also confirmed using the estimated natural logarithm (ln) of the probability of data in structure, which increased substantially from K=1 (ln=−9555.4) to K=4 (ln=−8822.5) and remained relatively stable thereafter. The Hex River Valley and Stellenbosch E. acerbella populations appeared closely related with the Elgin, Ceres and Tulbagh populations forming a second group (fig. 2). Bayesian analyses confirmed that the population structure of Thaumatotibia leucotreta populations could best be described by K=2 (ln=−17708.1). A similar pattern of population clustering was observed using neighbour-joining analysis, where two main clusters were observed, the first containing five of the seven Western Cape populations and the second containing the remaining Western Cape populations as well as all populations from Mpumalanga and the Eastern Cape (fig. 3).

Fig. 2. Cluster analysis (unrooted neighbour joining) showing the relationships between E. acerbella populations collected from seven regions in the Western Cape, based on AFLP analysis. Only bootstraps above 50% are shown.

Fig. 3. UPGMA cluster analysis based on analysis of 323 AFLP fragments showing the relationships among T. leucotreta populations collected from three different provinces in South Africa, using a laboratory colony maintained in Britain as an outgroup.
Gene flow among local populations
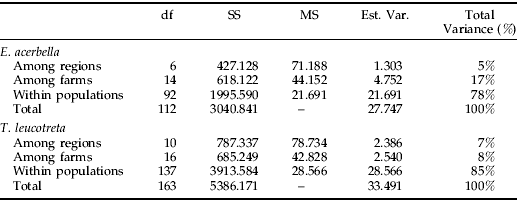
Analysis of molecular variance in E. acerbella populations indicated that, although the majority of genetic variability (78%, P=0.001) could be attributed to variation within populations, significantly high proportions could be ascribed to variation among the seven regions sampled (5%, P=0.001) as well as among different farms/sites (17%, P=0.001) (table 4). Similar patterns were detected in T. leucotreta populations, where AMOVA analysis showed that 7% (P=0.001) of the total variation could be attributed to variation among regions with a further 8% (P=0.001) of the variation due to among farms/sites (table 4). The remaining variation (85%, P=0.001) was due to individuals.
Table 4. Analysis of molecular variance (AMOVA), based on AFLP analysis, for E. acerbella and T. leucotreta populations collected in South Africa.
To further assess variation over local scales in E. acerbella, populations from three sites in Stellenbosch and 11 farms situated throughout the Hex River Valley were assessed. Evidence of population substructure was apparent in Stellenbosch populations, where 12% of the variation was attributed to variation among sites (ΦPT=0.121, P=0.001), as well as in Hex River Valley populations, where 22% of the variation was due to variation among farms (ΦPT=0.218, P=0.001; table 5). A similar experiment was conducted in T. leucotreta, using populations from seven farms in Citrusdal, three sites in Stellenbosch, three farms in the Sundays River Valley and four farms in Nelspruit. In Citrusdal, 8% of the total variation was as result of differences among farms (ΦPT=0.076, P=0.001; table 5). These values were calculated as 5% in Stellenbosch populations (ΦPT=0.055, P=0.001), 9% in Sundays River Valley populations (ΦPT=0.088, P=0.001) and 15% in Nelspruit populations (ΦPT=0.151, P=0.001). For both E. acerbella and T. leucotreta, individuals that were collected from the same site/farm often appeared to be closely related using cluster analysis; and, in some instances, it was possible, more or less, to ascribe individuals to populations based on their AFLP profiles. For example, cluster analysis illustrating relationships among E. acerbella individuals collected from the Hex River Valley is shown in fig. 4. Mantel tests confirmed that there was a low but significant correlation between genetic and geographic matrices for both species (r=0.18, P=0.001 in E. acerbella; and r=0.093, P=0.001 in T. leucotreta).

Fig. 4. Cluster analysis (UPGMA) showing the relationships between E. acerbella individuals collected from nine farms in the Hex River Valley, based on AFLP analysis. Bootstrap values above 50%, based on 1000 permutations, are indicated.
Table 5. AMOVA analysis for populations of E. acerbella and T. leucotreta collected on a local scale (within regions).

Gene flow among host populations
Estimates of population structure based on host populations were small but significant for both E. acerbella and T. leucotreta populations. Five percent of the variation in E. acerbella populations was due to hosts (ΦPT=0.050, P=0.001), whereas 8% was due to hosts in T. leucotreta populations (ΦPT=0.079, P=0.001) (table 6). However, for both species, estimates of population variation were not significantly higher when populations from more hosts were included in analyses at regional and local scales. In addition, it was not possible to distinguish among populations collected from different hosts using UPGMA analysis at any geographic scale. Bayesian analyses confirmed that hosts populations of both E. acerbella and T. leucotreta were best described by a single cluster (K=1, ln=−5912.7 and ln=−16074.8, respectively).
Table 6. Analysis of molecular variance of E. acerbella and T. leucotreta populations collected from various hosts.

Comparison among species
The origin and host status for each of the six tortricid species are shown in table 3, along with estimates of ΦPT and I. These parameters did not differ significantly at the species level when analyzed using a Chi squared test (P=1 for both parameters). Statistical differences between ΦPT and I values between native and introduced, as well as oligophagous and polyphagous, species were investigated using single factor ANOVA analysis. Estimates of ΦPT and I did not differ significantly when tested against population history (P=0.264 and 0.769, respectively). However, when investigating the effects of host range, ΦPT between oligophagous and polyphagous species were significantly different at less stringent levels (P=0.014) although I did not differ significantly (P=0.275).
Discussion
Gene flow among host populations
Little evidence was found to suggest that E. acerbella and T. leucotreta populations formed host strains, despite earlier suggestions that T. leucotreta races having different host preferences may exist (Ford, Reference Ford1934; Omer-Cooper, Reference Omer-Cooper1939). Thus, none of the six tortricids of major economic importance South Africa showed evidence of developing strains specific to hosts. The practical implication of these results is that uncultivated hosts may maintain populations at times when fruit is unavailable in the orchard, confirming suggestions that the proximity of other susceptible cultivated or wild fruits has a considerable influence on, for example, the severity of T. leucotreta infestation (Gunn, Reference Gunn1921; Daiber, Reference Daiber1981; Anderson, Reference Anderson1986). In addition, since populations maintained on uncultivated hosts may affect the efficiency of chemical control and the development of insecticide resistance by maintaining reservoirs of susceptible populations, pest management programs should take into account the presence of alternative hosts. It should be emphasized, however, that the results found for these tortricids in South Africa may not necessarily apply to these species where they occur in other countries. For example, C. pomonella host strains have been found to be specific to apple, apricots and walnuts in central Europe (Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008; Chen & Dorn, Reference Chen and Dorn2009).
Gene flow among geographic populations
All six tortricid species examined, including E. acerbella and T. leucotreta, showed evidence of being structured geographically. In addition, using AFLP analysis, it was possible to distinguish between some populations collected from the same region as well as, in certain instances, different farms or even orchards. Thus, it appears as if gene flow among populations of each of the six species is limited on a local scale. A number of factors may counteract the effects of gene flow and produce genetic structure over geographic scales in populations of phytophagous insect pests. Here, we consider the impact of host range, origin, dispersal ability, control practices and anthropogenic movement on the population genetic structure of the six tortricid species analyzed.
Host range
It seems instinctive to assume that polyphagous species will display relatively pronounced levels of population subdivision, given their relative abundance of hosts that may act as staging posts for dispersal and subsequent gene flow. However, estimates of population subdivision for polyphagous C. peltastica, E. acerbella and T. leucotreta were significantly different to oligophagous C. pomonella, G. molesta and T. batrachopa only at less stringent significance levels. These results seem to confirm studies, such as that by Peterson & Denno (Reference Peterson and Denno1998), that diet breadth alone cannot be used to infer the population genetic structure of a species. Host range, thus, seems to have played a relatively small role in determining the population genetic structure of economically important Tortricidae in South Africa.
Historical effects
Historical effects, including the introduction of pest species to regions where they are not native, have long been known to affect population genetic structure. The tortricids E. acerbella, T. leucotreta, C. peltastica and T. batrachopa are native to southern Africa (Myburgh & Basson, Reference Myburgh and Basson1961; Oliver & Bolton, Reference Oliver and Bolton1974; Quilici et al., Reference Quilici, Verbizier, Trahais and Manikom1988; Newton, Reference Newton, Bedford, van den Berg and de Villiers1998). In contrast, C. pomonella was first reported in South Africa in 1885 (Lounsbury, Reference Lounsbury1898; Giliomee & Riedl, Reference Giliomee and Riedl1998), whereas G. molesta was first observed only in 1990, but may have been present earlier (Blomefield & Geertsema, Reference Blomefield and Geertsema1990; Timm et al., Reference Timm, Geertsema and Warnich2008). Because of the recent introduction of C. pomonella and G. molesta, it might be expected that their populations are likely to be much less genetically diverse than the populations from which they are derived (Barrett & Kohn, Reference Barrett, Kohn, Falk and Holsinger1991). However, both species displayed similar levels of diversity to that of native species. In addition, patterns of C. pomonella genetic structure in South Africa are comparable to that in its native range in Europe (Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008). It is likely that both C. pomonella and G. molesta, which are known to be successful invaders, were introduced into South Africa multiple times (Blomefield & Geertsema, Reference Blomefield and Geertsema1990; Giliomee & Riedl, Reference Giliomee and Riedl1998; Timm et al., Reference Timm, Geertsema and Warnich2008), which may have contributed to its genetic diversity (Sakai et al., Reference Sakai, Allendorf, Holt, Lodge, Molofsky, With, Baughman, Cabin, Cohen, Ellstrand, McCauley, O'Neil, Parker, Thompson and Weller2001). Because of their recent history in South Africa, both species may not be in mutation-drift equilibrium, and it is possible that effects observed are more relevant to historical patterns of gene flow than to the current population dynamics (Avise, Reference Avise1994; Bossart & Pashley Prowell, Reference Bossart and Pashley Prowell1998). However, since the other four species analysed all produced similar patterns of gene flow and C. pomonella is known to display similar patterns in its native range (Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008; Chen & Dorn, Reference Chen and Dorn2009), it is likely that the results found in South Africa could be relevant elsewhere.
Dispersal ability
Limited dispersal ability may be a distinguishing feature of the six Tortricidae analysed in South Africa. AFLP analysis was able to distinguish between closely situated populations of both E. acerbella and T. leucotreta, indicating that local strains have evolved within regions in these species. These results are similar to those produced for C. pomonella (Timm et al., 2006a; Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008; Chen & Dorn, Reference Chen and Dorn2009), C. peltastica and T. batrachopa (Timm et al., Reference Timm, Geertsema and Warnich2006b) and G. molesta (Timm et al., Reference Timm, Geertsema and Warnich2008). Mark-recapture or flight-mill studies of C. pomonella, G. molesta and T. leucotreta have shown that, generally, individuals vary greatly in their dispersal capacity (Mani & Wildbolz, Reference Mani and Wildbolz1977; Sziraki, Reference Sziraki1979; Vickers et al., Reference Vickers, Rothschild and Jones1985; Rothschild & Vickers, Reference Rothschild, Vickers, Van der Geest and Evenhuis1991; Schumacher et al., Reference Schumacher, Weber, Hagger and Dorn1997a,Reference Schumacher, Weyeneth, Weber and Dornb; Newton, Reference Newton, Bedford, van den Berg and de Villiers1998; Keil et al., Reference Keil, Gu and Dorn2001; Gu et al., Reference Gu, Hughes and Dorn2006). Although individuals may undertake single long flights of up to several kilometres, the majority of individuals appear to be fairly sedentary. In C. pomonella, there is known to be a trade-off between mobility and fitness, which may favor sedentary individuals (Gu et al., Reference Gu, Hughes and Dorn2006). Variation in dispersal has previously been explained in terms of the life history of tortricid moths in an orchard (Schumacher et al., Reference Schumacher, Weber, Hagger and Dorn1997a; Timm et al., Reference Timm, Geertsema and Warnich2006a). If sufficient resources are readily available, it may be advantageous for moths to stay within the habitat or the orchard. However, individuals with the capacity for a longer flight range will be favoured when the food resource of the larvae fluctuates. It is most likely that such individuals, infesting newly-established orchards, cause subsequent patterns of variation since moths in newly established orchards appear to be more closely related to each other than to moths from established orchards. The sedentary nature of individuals, thus, may favour splitting into many local populations, aided by the effects of genetic drift.
Insecticide use
Conventional chemical treatments may also be responsible for producing locally adapted populations. This pattern was highlighted in C. pomonella populations in France, where populations were found to be mainly structured according to the history of insecticide applications (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007). It may be likely that a similar effect is evident in South African populations, since chemical sprays form a major component of pest management programs against tortricids in South Africa (Newton, Reference Newton1989; Blomefield, Reference Blomefield1994, Reference Blomefield1996; Hofmeyr & Pringle, Reference Hofmeyr and Pringle1998; Riedl et al., Reference Riedl, Blomefield and Giliomee1998; Blomefield & Barnes, Reference Blomefield and Barnes2000; Blomefield & du Plessis, Reference Blomefield and Barnes2000).
Anthropogenic movement
Anthropogenic movement of fruit or other plant material also may have affected the population genetic structure of the Tortricidae analysed (Higbee et al., Reference Higbee, Calkins and Temple2001; Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007). This effect may be evident in the close relationship between E. acerbella populations collected from the Hex River Valley and Stellenbosch, as both regions are known for intensive wine grape production, which may have facilitated the exchange of plant material between regions. Patterns of population variation between regions in T. leucotreta may have been similarly affected by agricultural practices. For example, it has been suggested that the Western Cape population has its origin from material originally obtained from Mpumalanga province (Giliomee & Riedl, Reference Giliomee and Riedl1998), which could possibly explain the close relationships between the Elgin and Nelspruit populations.
Conclusion
The Tortricidae, of major economic importance in South Africa, appear to be structured genetically as a result of geography and, to a lesser extent, host use. Furthermore, populations are structured over local spatial scales, most likely as result of limited dispersal ability, insecticide use and anthropogenic movement. The aforementioned factors can be manipulated and, therefore, should be taken into account in a pest management program. For example, various pest management practices, such as the release of parasitoids or sterilized insects and the placement of insecticide treatments, are affected by limited dispersal ability. The results produced, thus, are useful for understanding the ecology of the Tortricidae of economic importance in South Africa, facilitating in turn an improvement of their eventual control.












