Introduction
Helicoverpa is an important lepidopteran genus containing 18 species, of which four species, H. armigera (Hübner), H. zea (Boddie), H. punctigera (Wallengren) and H. assulta (Guenée), are significant agricultural pests and collectively cause economic losses in billions of US dollars annually. Other species in this group can be pests, but they are either greatly limited in host plant range or in geographic distribution. H. armigera has the widest distribution, occurring throughout Asia, Africa, Europe and Australasia. The very closely related H. zea (Mitter et al., Reference Mitter, Poole and Matthews1993; Behere et al., Reference Behere, Tay, Russell, Heckel, Appleton, Kranthi and Batterham2007) occurs in North and South America, while H. punctigera is endemic to Australia. Species identification based on external morphology of these noctuid moths is difficult, even using forewing morphology (Matthews, Reference Matthews1999). Evolutionary development within the Helicoverpa genus is more readily seen in changes to the morphology of genitalia (Hardwick, Reference Hardwick1965). Various molecular and biochemical methods have been proposed for species diagnosis in this genus. H. armigera can be differentiated from H. punctigera based on a DNA-PCR of the ITS region (Pearce, Reference Pearce2003), allozyme electrophoresis (Daly & Gregg, Reference Daly and Gregg1985) and immunoassay (Trowell et al., Reference Trowell, Forrester, Garsia, Lang, Bird, Hill, Skerritt and Daly2000). Distinguishing between H. assulta and H. armigera is possible based on PCR-RFLP (Kranthi et al., Reference Kranthi, Kranthi, Bharose and Syed2005) and AFLP markers (Ming & Wang, Reference Ming and Wang2006). Monoclonal antibodies raised against H. zea can distinguish, inter alia, H. armigera from H. punctigera (Greenstone et al., Reference Greenstone, Stuart and Haunerland1991). However, currently there are no uniform molecular DNA techniques that can easily separate the four pest species from each other (i.e. H. zea from H. armigera and H. assulta, or H. assulta from H. Punctigera).
Here, we report on a life stage-independent PCR-RFLP molecular DNA technique based on two partial regions of the mitochondrial DNA (mtDNA) genome for distinguishing between H. armigera, H. zea, H. punctigera and H. assulta.
Materials and methods
Samples and DNA extraction
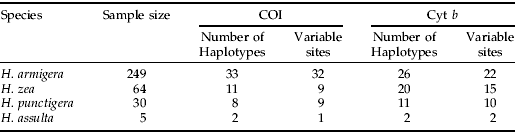
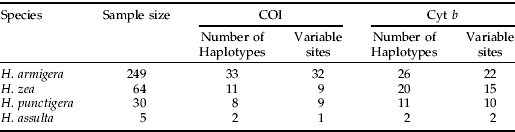
Samples of H. armigera (n=249), H. zea (n=64), H. punctigera (n=30) and H. assulta (n=5) were field-collected from various continents. Collection details and DNA extraction protocols are as described in Behere et al. (Reference Behere, Tay, Russell, Heckel, Appleton, Kranthi and Batterham2007). Briefly, H. armigera were obtained from Australia (n=56; larvae and moths), China (n=34; larvae), Burkina Faso (n=35; larvae), Uganda (n=24; larvae), India (n=90; larvae, pupae and moths) and Pakistan (n=10; larvae). H. punctigera were collected from Australia and H. assulta were collected from India. H. zea were from the USA (n=34; larvae and moths) and Brazil (n=30; moths). H. punctigera and H. assulta were collected as moths and larvae, respectively. Additionally, eggs from laboratory-reared H. armigera (collected from Australia) at different stages of maturation (one to four days old) were placed individually in 1.7 ml sterile Eppendorf tubes, and DNA was extracted using the DNeasy® Tissue Kit (Qiagen, Cat. No 69504). DNA from eggs was eluted in 20 μl elution buffer and stored at −20°C until needed.
PCR and sequencing
PCR and sequencing primers for cytochrome oxidase I (COI) and cytochrome b (Cyt b) were designed from GeneBank sequences (accession numbers, AY437834, AY437835, DQ059302 and AF467260) using the primer designing program Oligo 6.4 (Molecular Biology Insights, Inc.). Two mtDNA primer sets were designed for COI and Cyt b. The primer set COI-F02 (5′ CTC AAA TTA ATT ACT CCC CAT C 3′) and COI-R02 (5′ GGA GGT AAG TTT TGG TAT CAT T 3′) was used to amplify 511 base pairs (bp) of partial COI. The primer set Cytb-F02 (5′ GAA TCC TTT AAT TTA AAA TAT AC 3′) and Cytb-R02 (5′ AAA TAT GGG TTA GTT AAA GTT AA 3′) was used to amplify 434 bp of partial Cyt b. PCR conditions had the following profile: 94°C for 4 min (one cycle), 30 s each of 94°C, 50°C and 1 min at 72°C (35 cycles) followed by a final extension cycle of 72°C for 10 min. The proof of DNA amplification was confirmed by running out 5 μl of the post-PCR volume on 1.5% agarose gels. PCR amplification of individual DNA samples was carried out in a total of 25 μl reaction volume and contained approximately 25 ng of genomic DNA, 0.2 μm each of forward and reverse primers, 0.2 mm of dNTP's, 1×PCR reaction buffer (Promega), 1.5 mm Mg2+ and one unit of Taq DNA polymerase (Promega). We directly sequenced all 348 individuals for Cyt b from both ends for accuracy. The sequencing protocol was as reported in Behere et al. (Reference Behere, Tay, Russell, Heckel, Appleton, Kranthi and Batterham2007). COI sequences (GenBank Accession numbers: EF116226-EF116274) of all samples were as reported in Behere et al. (Reference Behere, Tay, Russell, Heckel, Appleton, Kranthi and Batterham2007) with the exception of H. punctigera (accession numbers EF410014-EF410019) because these represented additional six haplotypes from 25 new samples detected in this study. All Cyt b nucleotide sequences obtained were submitted to the National Centre for Biotechnology Information (NCBI; GenBank Accession numbers EF410020–EF410078). Partial COI and Cyt b DNA fragments of the H. armigera egg samples were not sequenced.
Restriction fragment length polymorphism analysis
Restriction sites were predicted from the COI and Cyt b sequence data using NEBcutter V2.0 <http://tools.neb.com/NEBcutter2/index.php>. For each sample, 5 μl of PCR product was digested with two units of restriction enzyme (BstZ17I or HphI) in a 20 μl reaction volume according to the manufacture's instructions (New England Biolabs). Following incubation at 37°C for 6 h, the digested products were separated by electrophoresis on 2.5% 1×TAE agarose gels at 180V for 30 min and stained with ethidium bromide. In order to confirm restriction band sizes accurately, the digested products were also tested on 6% polyacrylamide gels. The size of restriction fragments was estimated using a Kodak® Electrophoresis Documentation and Analysis System (EDAS 290) by comparison with the BenchTop pGEM® DNA Ladder molecular weight standard (Promega).
Results
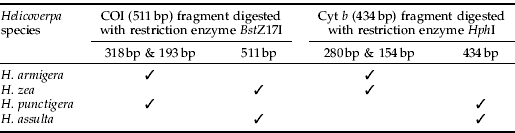
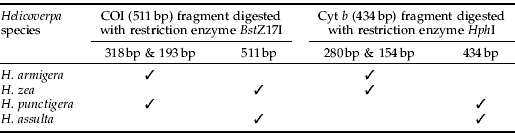
A total of 348 individuals from the four agriculturally significant Helicoverpa pest species were sequence-analysed. For each individual, partial COI (511 bp) and Cyt b (434 bp) genes were obtained from PCR amplification. DNA from all individuals tested in this study was amplified successfully, including the DNA extracted from single eggs. The numbers of haplotypes and of variable sites identified at COI and Cyt b in all species are presented in table 1. Within partial COI sequences, a single base pair mutation was found at the BstZ17I recognition site (GAATAC) in H. zea and H. assulta but not in H. armigera and H. punctigera (GTATAC). Restriction endonuclease digest of the 511 bp COI PCR product, therefore, gave two bands of 318 bp and 193 bp in H. armigera and H. punctigera but not in H. zea and H. assulta (fig. 1).

Fig. 1. Partial COI (511 bp) RFLP pattern (digested with BstZ17I) of H. armigera, H. zea, H. punctigera and H. assulta, with bands separated on 6% polyacrylamide gel. Lanes labeled as MS are pGEM molecular weight standard DNA ladders.
Table 1. Number of haplotypes and variable sites detected at partial COI (511 bp) and Cyt b (434 bp) in the four Helicoverpa pest species.
Within the 434 bp of Cyt b sequences from the four pest species, the recognition site for HphI (GGTGA (N)8) was present in H. armigera and H. zea but absent in H. assulta (GATGG) and H. punctigera (GATGT). Restriction digestion of the 434 bp Cyt b PCR product, therefore, resulted in two bands of 280 bp and 154 bp in H. armigera and H. zea (fig. 2, table 2). Restriction banding patterns generated were easily visualised on 2.5% agarose gels. Digested PCR amplicons fragment sizes estimated from 6% polyacrylamide gels were also confirmed by DNA sequencing data.

Fig. 2. Partial Cyt b (434 bp) gene amplified from H. armigera, H. zea, H. punctigera and H. assulta and restriction digested with Hph I, with bands separated on 6% polyacrylamide gel. Lanes labeled as MS are pGEM molecular weight standard DNA ladders.
Table 2. Restriction fragment length polymorphism (RFLP) patterns at COI (511 bp) and Cyt b (434 bp) in pest species of Helicoverpa.
Discussion
Species separation based on PCR-RFLP patterns at a different partial mtDNA COI region has previously been demonstrated for H. armigera and H. assulta (Kranthi et al., Reference Kranthi, Kranthi, Bharose and Syed2005), although the sample sizes tested for H. armigera and H. assulta were smaller than in this study. In China, AFLP markers have been proposed for identification of these two species (Ming & Wang, Reference Ming and Wang2006). Combining restriction digest patterns of partial mtDNA COI and Cyt b genes with the two endonucleases allowed differentiation of the four pests Helicoverpa species and the tested life stages (egg, larva, pupae and adults) in H. armigera. We have not specifically tested all four life stages in the other three Helicoverpa pest species, although results from H. armigera indicated this to unlikely represent significant problems.
The accuracy of RFLP band sizes was initially determined on 6% polyacrylamide gel electrophoresis (PAGE) and confirmed by sequencing data. All remaining samples were screened on 2.5% agarose gels. Although 6% PAGE provided better resolution for smaller fragments (i.e. less than 200 bp), (figs 1 and 2), we were able to easily score the 154 bp fragment in H. armigera and H. zea as digested by HphI on 2.5% agarose gels (figures not shown). For routine identification, the use of agarose gels is preferred over PAGE, as this considerably reduces chemical and reagent costs and is non-toxic. The PCR amplification protocol and primer sets described in this study are very robust, as have been demonstrated in cross-species PCR amplification. Although PCR amplification can facilitate the use of minute quantities of DNA template from samples that have not been optimally preserved (Taylor & Szalanski, Reference Taylor and Szalanski1999), we recommend only using well-preserved samples (i.e. consistently stored at between −20°C to −70°C or kept in a sufficient volume of ≥95% ethanol) in PCR-RFLP analyses of these four pest Helicoverpa species. The procedures described in this study, from DNA extraction, digestion, gel electrophoresis to species identification, can be accomplished within a single working day. The use of more than one restriction endonuclease will further minimise species misidentification.
In this study, we have sequenced only five H. assulta individuals and identified only two mtDNA haplotypes. The PCR-RFLP patterns for H. armigera, H. zea and H. punctigera based on the partial COI and Cyt b gene regions were clearly distinguishable from those of H. assulta, while phylogenetic analysis based on the COI region (Behere et al., Reference Behere, Tay, Russell, Heckel, Appleton, Kranthi and Batterham2007) clustered the two H. assulta mtDNA haplotypes together after 1000 bootstrap re-sampling analyses with a very high support value (98%). Nevertheless, it would be desirable to increase our H. assulta sample size, especially from other parts of Asia (e.g. China) and from Australia and Africa. Performing a nucleotide–nucleotide BLAST search (BLASTN: Altschul et al., Reference Altschul, Madden, Schaffer, Zhang, Zhang, Miller and Lipman1997), using the two H. assulta mtDNA COI haplotypes detected in this study (EF116270, EF116271) against the non-redundant (nr) DNA database, showed no overlapping with the H. assulta partial COI sequence (AY264943) submitted by Kranthi et al. (Reference Kranthi, Kranthi, Bharose and Syed2005) at the BstZ17I restriction enzyme recognition site; while the H. armigera BstZ17I restriction site identified in this study from all 33 haplotypes further matched an additional H. armigera sequence available in GenBank (AY437834).
Given the differential response in tolerance to insecticides among these pest species, the ability to distinguish pest species at the egg and early instar larval life-stages has important implication for pest management because insecticides are usually most effective at these life-stages. Air-freight transportation of agricultural commodities between continents is an ever-increasing activity, especially with vegetables and ornamentals. Almost every year, H. zea caterpillars are intercepted on agricultural and horticultural produce to the UK (Seymour, Reference Seymour1978). Regular interception of H. armigera or Helicoverpa species in the USA have been commonly reported, with Venette et al. (Reference Venette, Davis, Zaspel, Heisler and Larson2003) reporting 4431 interceptions in the USA since 1985 on fruits, vegetables, ornamentals and other miscellaneous plants and Pouge (Reference Pouge2004) adding a further 20. The risk assessment study conducted by Venette et al. (Reference Venette, Davis, Zaspel, Heisler and Larson2003) gave a high potential entry and establishment rating for H. armigera into the North American continent. Due to the extreme similarity of H. zea and H. armigera at larval stages, it is extremely difficult to separate these pest species without rearing larvae to adulthood followed by examining genitalia of adults through dissection (<http://www.eppo.org/QUARANTINE/insects/Helicoverpa_zea/HELIZE_ds.pdf>; EPPO data sheets on Quarantine Pests, Helicoverpa zea). For quarantine purposes, PCR primers must faithfully amplify samples stored under a variety of conditions. The mtDNA diagnostic primers reported in this study have been designed, based on highly conserved mtDNA regions, to amplify short fragments of COI and Cyt b genes and, hence, to take advantage of greater efficiencies of PCR at amplification of short DNA fragments (Muraji & Nakahara, Reference Muraji and Nakahara2002).
Acknowledgements
We would like to thank all the people who kindly provided samples used in this study, particularly Keshav Kranthi (India), Sandhya Kranthi (India), David Heckel (Germany), A. Regupathy (India), Mushtaq Ahmed (Pakistan), Yidong Wu (China), Philippe Menozzi (Burkina Faso), Peter Ridland (Australia), Nancy Endersby (Australia), Stephen Cameron (USA), Dan Gilrein (USA) and Pierre Slivie (Brazil). H. armigera egg samples were kindly provided by Adam Williams (The University of Melbourne). Mark Blacket (The University of Melbourne) assisted with polyacrylamide gel electrophoresis. This project was supported by grants from the Melbourne International Research Scholarship (MIRS) and Melbourne International Fee remission Scholarship (MIFRS) to GTB. WTT was supported by grants from the State Government of Victoria and the Max-Planck-Gesellschaft. This work was also supported by the Australian Research Council (ARC) through its funding to the Special Research Centre for Environmental Stress and Adaptation Research (CESAR).