Introduction
Chagas disease is a life-threatening disease caused by the vector-transmitted protozoan parasite Trypanosoma cruzi, affecting approximately 7 million people worldwide, mostly in Latin American countries, and causing over 10 000 deaths per year (World Health Organization, 2017). In the past few years, the disease has spread to other countries due to migration and blood transfusion. During its complex life cycle, the parasite alternates between the insect vector and a mammalian host, with at least four distinct well-characterized life-cycle forms: replicative epimastigotes that differentiate into infective metacyclic trypomastigotes as the parasites move along the digestive tract of the insect vector (metacyclogenesis) and replicative amastigotes (intracellular forms) and infective bloodstream trypomastigotes in mammalian hosts (Brener, Reference Brener1973).
The different life cycle stages of T. cruzi are faced with distinct hostile environments (Rodrigues et al., Reference Rodrigues, Godinho and de Souza2014). Survival and infectivity of the parasite rely on cell-surface and secreted glycoconjugates, some of which display unique carbohydrate structures. Glycoinositolphospholipids (GIPLs) and highly abundant glycoproteins, encoded by multigene families, such as mucins, mucin-associated surface proteins (MASPs) and trans-sialidases (TSs), display stage-specific gene expression and are critical virulence factors, involved in the protection against proteases present in the insect vector and in the invasion and modulation of the host immune system (Buscaglia et al., Reference Buscaglia, Campo, Frasch and Di Noia2006; de Lederkremer and Agusti, Reference de Lederkremer and Agusti2009; De Pablos and Osuna, Reference De Pablos and Osuna2012; Mucci et al., Reference Mucci, Lantos, Buscaglia, Leguizamón and Campetella2017).
Many of T. cruzi glycoconjugates, including GIPLs, mucins and the gp72 protein, which contains a unique and complex P-linked glycan (Allen et al., Reference Allen, Richardson, Mehlert and Ferguson2013), are rich in galactose residues, either in the form of galactopyranose (Galp) or in galactofuranose (Galf) (de Lederkremer and Agusti, Reference de Lederkremer and Agusti2009). In particular, β-Galp residues present in O-linked oligosaccharides in T. cruzi mucins are the primary acceptors for sialylation catalyzed by the parasite-unique trans-sialidase (Schenkman et al., Reference Schenkman, Ferguson, Heise, de Almeida, Mortara and Yoshida1993), whose activity is essential for parasite survival and infectivity (Dc-Rubin and Schenkman, Reference Dc-Rubin and Schenkman2012). Furthermore, α-Galp residues, which are also present in T. cruzi mucins, are strongly immunogenic, representing the main target of acute (Gazzinelli et al., Reference Gazzinelli, Pereira, Romanha, Gazzinelli and Brener1991) and chronic (Almeida et al., Reference Almeida, Milani, Gorin and Travassos1991, Reference Almeida, Ferguson, Schenkman and Travassos1994) Chagas disease antibodies.
The importance of galactose metabolism in T. cruzi was demonstrated by the knockout of the TcGALE gene coding for UDP-glucose 4′-epimerase, responsible for the epimerization of UDP-glucose to UDP-galactose (MacRae et al., Reference MacRae, Obado, Turnock, Roper, Kierans, Kelly and Ferguson2006). This is the only source of galactose in the parasite since its hexose transporter system is unable to transport galactose (de Lederkremer and Agusti, Reference de Lederkremer and Agusti2009). TcGALE is an essential gene, and single knockout mutants, in which only one allele has been knocked out, display biochemical and morphological alterations and a strong reduction in the expression of galactose-containing mucins (MacRae et al., Reference MacRae, Obado, Turnock, Roper, Kierans, Kelly and Ferguson2006). The addition of galactose residues to T. cruzi glycoconjugates occurs in the lumen of the Golgi apparatus through the action of glycosyltransferases using nucleotide sugars as substrates. These activated sugar donors must be transported from the cytosol into the Golgi lumen by nucleotide-sugar transporters (NSTs). These transporters have been characterized in different organisms, and their essential role in glycosylation has been demonstrated in mammals, yeasts, plants, Caenorhabditis elegans and protozoan parasites (Caffaro and Hirschberg, Reference Caffaro and Hirschberg2006; Liu and Hirschberg, Reference Liu and Hirschberg2012).
Recently, we identified an UDP-N-acetylglucosamine (UDP-GlcNAc) transporter in T. cruzi (TcNST1), which is essential for T. cruzi survival (unpublished results). In this work, we identified an UDP-galactose (UDP-Gal) transporter in T. cruzi by complementation of mutant mammalian Chinese Hamster Ovary (CHO) cells. This transporter, named TcNST2, is localized to the Golgi apparatus and seems to be expressed throughout the life cycle of the parasite. Due to the importance of galactose for T. cruzi and the other two medically relevant trypanosomatids, T. brucei spp and Leishmania spp. (Turnock and Ferguson, Reference Turnock and Ferguson2007), TcNST2 may play an important role in the synthesis of glycoconjugates and, therefore, in parasite survival and infectivity.
Materials and methods
Cell culture
Epimastigotes of the Trypanosoma cruzi Dm28c clone were grown in liver-infusion tryptose (LIT) medium supplemented with 10% fetal bovine serum (FBS) at 28 °C, as previously described (Camargo, 1964). Epimastigotes in exponential growth were transfected by electroporation (Lu and Buck, Reference Lu and Buck1991), and selection of stable transfectants was performed with G418 (500 µg mL−1). Metacyclic trypomastigotes were obtained by in vitro differentiation of epimastigotes as described (Contreras et al., Reference Contreras, Salles, Thomas, Morel and Goldenberg1985). Cell-derived trypomastigotes were obtained from the supernatant of 4-day-old infected VERO cells by centrifugation. Axenic amastigotes were obtained by cell-derived trypomastigotes submitted to low pH for 48 h (Tomlinson et al., Reference Tomlinson, Vandekerckhove, Frevert and Nussenzweig1995).
Chinese hamster ovary cell lines CRL-1781 (Stanley, Reference Stanley1981) and Lec8, deficient in UDP-Gal transport (Deutscher and Hirschberg, Reference Deutscher and Hirschberg1986), were purchased from the American Type Culture Collection (ATCC) and maintained in RPMI medium with L-glutamine supplemented with 10% FBS, 100 UI mL−1 penicillin, 10 µg mL−1 streptomycin and 2 mm glutamine at 37 °C in an atmosphere of 5% CO2. CHO Lec8 cells were transfected with BglII-linearized plasmids containing the putative T. cruzi NST genes (see details in Plasmid construction) using Lipofectamine 2000 (Invitrogen). Stable transfectants were obtained after serial passages in presence of 1 mg mL−1 G418 until untransfected control cells died. VERO cells were maintained in high glucose DMEM medium containing the same supplements as CHO cells.
Complementation assays
Mutant CHO Lec8 cells stably expressing putative T. cruzi UDP-Gal transporters, as described above, were harvested by trypsinization, fixed with 1% formaldehyde and incubated with GS-II lectin Alexa Fluor 488 conjugate (Molecular Probes, Inc.) in PBS (40 μg mL−1) under gentle agitation for one hour at room temperature. Cells were then washed twice with PBS and lectin binding was evaluated by flow cytometry in a BD FACSCanto™ II (BD Biosciences, USA). Fluorescence data were analyzed with FlowJo software (FlowJo, Ashland, OR, USA, version 7.7).
For microscopic analysis, wild-type and mutant CHO cells were plated on coverslips and cultured for 4 days. Cells were fixed with 4% paraformaldehyde for one hour at room temperature, blocked by incubation overnight with 3% BSA and then incubated with GS-II lectin Alexa Fluor 488 conjugate (40 µg mL−1) (Molecular Probes, Inc.) and Hoechst 33258 DNA dye (1 µg µL−1) (Thermo Scientific) for one hour at room temperature. After extensive washing, the coverslips were mounted onto glass microscope slides using ProLong® Gold antifade (Thermo Scientific) and examined using a Leica DMI6000 B inverted microscope (Leica Microsystems, Mannheim, Germany).
Plasmid construction
For the complementation assays in mutant CHO Lec8 cells, the coding sequences of the selected genes (TcCLB.504085.60, TcCLB.511277.400 and TcCLB.506509.40, according to the TriTrpDB database) were amplified by PCR from T. cruzi genomic DNA with the high-fidelity polymerase Pfx (Invitrogen) using specific primers containing appropriate restriction sites. After digestion with XhoI/EcoRI (TcCLB.504085.60 and TcCLB.506509.40) or EcoRI (TcCLB.511277.400), the PCR products were cloned into the pcDNA3.1 (-) vector (Invitrogen). The endogenous CHO UDP-Gal transporter was cloned into the same vector and used as a positive control. The corresponding gene was RT-PCR-amplified from total RNA samples using Superscript III Reverse Transcriptase (Invitrogen) followed by amplification of the cDNA with Pfx. RNA samples were obtained from wild-type CHO cells (CRL-1781 strain) with the NucleoSpin RNA Kit (Macherey-Nagel). A summary of the primers and the restriction sites used for cloning are indicated in Supplementary Table S1.
For expression in T. cruzi, TcNST2 was cloned into the pDONR221 vector (Invitrogen) and then transferred by recombination to the destination vector pTcGFPN (Batista et al., Reference Batista, Marchini, Celedon, Fragoso, Probst, Preti, Ozaki, Buck, Goldenberg and Krieger2010) in frame with Green Fluorescence Protein (GFP) at the carboxy-terminal (TcNST2-GFP).
Immunofluorescence analysis
Transfected epimastigotes expressing TcNST2-GFP were grown to an exponential phase, harvested by centrifugation, fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100. After blocking with 2% BSA, cells were incubated with rabbit anti-GFP (1:500) and mouse anti-TcHIP (1:250), in 2% BSA – TcHIP is a putative T. cruzi zDHHC palmitoyl transferase, used as a Golgi marker (Batista et al., Reference Batista, Kalb, Moreira, Batista, Eger and Soares2013). Goat anti-rabbit IgG (Alexa Fluor 488) and goat anti-mouse IgG (Alexa Fluor 546) (Molecular Probes) were used as secondary antibodies. Slides were examined using a Leica confocal microscope. Polyclonal antibodies were produced in mice according to international guidelines for the ethical use of animals.
RNA isolation and qPCR
Total RNA was isolated from the four life-cycle forms of T. cruzi (two independent samples for epimastigotes and cell-derived trypomastigotes and three samples for metacyclic trypomastigotes and axenic amastigotes) using the NucleoSpin RNA Kit (Macherey-Nagel). Any contaminant DNA was further digested in solution with the DNase provided by the kit as recommended by the manufacturer. Aliquots of 500 ng of total RNA were then reverse transcribed to first strand cDNA using oligo (dT) primers and Superscript III Reverse Transcriptase (Invitrogen) at 50 °C for an hour followed by inactivation at 70 °C for 15 min. Controls without the reverse transcriptase were performed for each RNA sample. qPCR assays were performed in triplicate with the SYBR Select Master Mix (Applied Biosystem) at the recommended concentration and 200 nm of specific primers using the LightCycler System (Roche). A five-point standard curve of 1:10 dilution was performed in triplicate to assess primer efficiency for the target gene (TcNTS2) and reference genes (histone 2B and the 60S ribosomal protein L9). Following amplification, a melting temperature analysis was carried out with a temperature increase from 65 to 95 °C at 0.1 °C s−1 to investigate the product's specificity. Data analysis was performed using the Pfaffl model (Pfaffl, Reference Pfaffl2001). After adjusting the Cq values according to primer efficiency, the stage/reference ratios were calculated separately for L9 and histone 2B. The final normalized expression ratios were then calculated by the average of the normalized ratios obtained for each sample and reference gene.
Structure prediction and analysis
A three-dimensional model of T. cruzi TcNST2 was generated using the I-TASSER server (Yang et al., Reference Yang, Yan, Roy, Xu, Poisson and Zhang2015). Structural alignment with the Saccharomyces cerevisiae GDP–mannose (GDP-Man) transporter Vrg4 (PDB 5OGE and 5OGK) was performed using TM-align (Zhang and Skolnick, Reference Zhang and Skolnick2005).
Statistical analysis
For the median fluorescence data in complementation assays and qPCR data, the statistical analysis was performed using one-way ANOVA followed by Dunnett's multiple comparisons test. Graphs were constructed using GraphPad Prism Software (GraphPad Prism, Inc., San Diego, CA, USA, version 6.01).
Results
T. cruzi UDP-Gal transporter
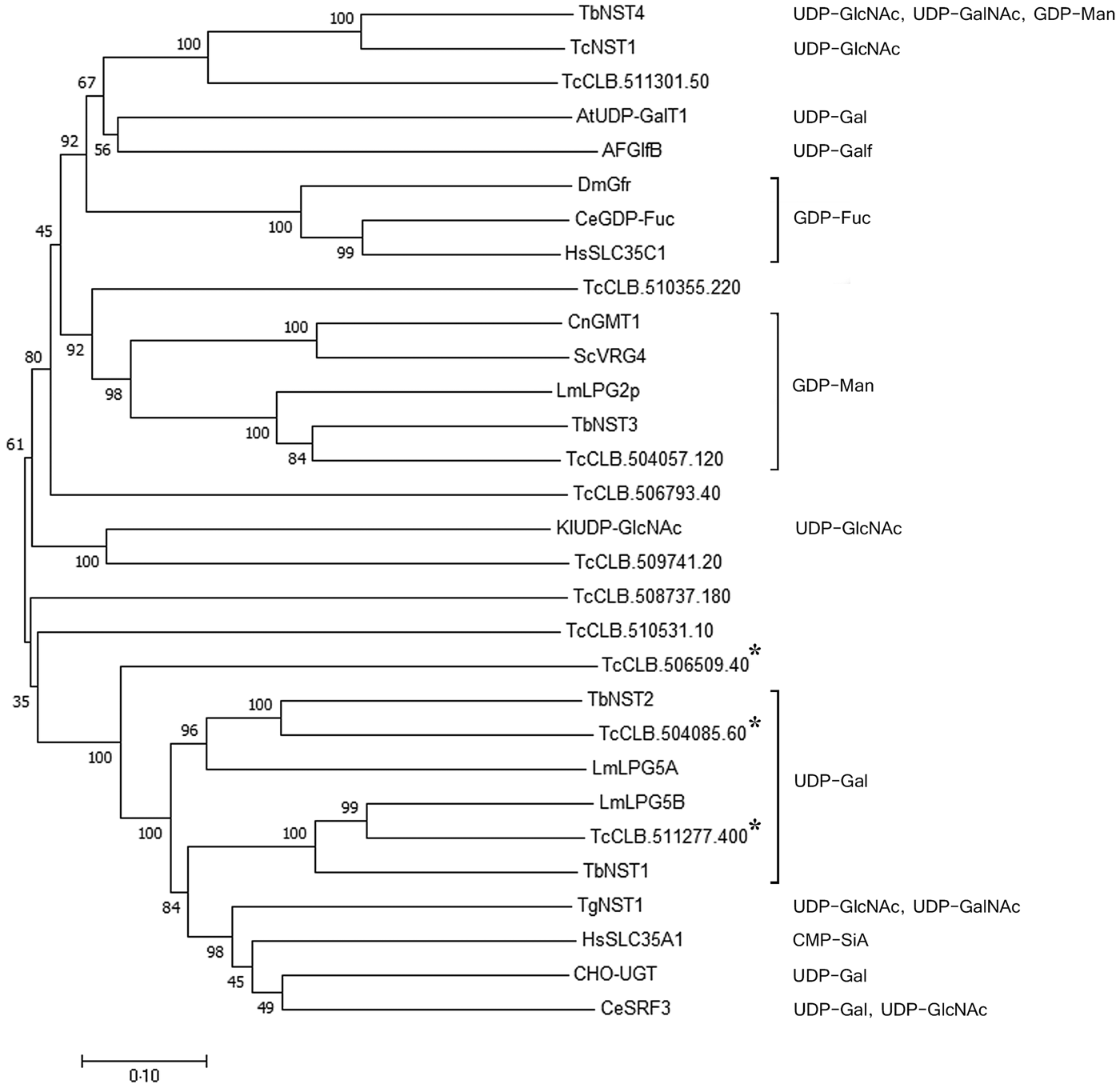
Previously, we identified a family of eleven putative nucleotide sugar transporters in the T. cruzi genome (Baptista et al., Reference Baptista, Rodrigues, Morking, Klinke, Zardo, Soares, de Aguiar, Goldenberg and Ramos2015). The prediction of substrate specificity for NSTs based on sequence analysis, however, is unreliable, especially for UDP-sugars. A phylogenetic analysis using experimentally characterized NSTs from other organisms and the T. cruzi NST candidates exemplifies this observation (Fig. 1). GDP-Man and GDP-fucose (GDP-Fuc) transporters from different organisms are clustered together, but the UDP-sugars clustering due to substrate specificity is less clear. UDP-Gal transporters are found in different clades and clustered not only together but also with transporters for UDP-GlcNAc, UDP-N-acetylgalactosamine (UDP-GalNAc) and CMP-sialic acid (CMP-Sia). Nevertheless, UDP-Gal transporters from Leishmania major (LPG5A and LPG5B) and T. brucei (TbNST1 and TbNST2) form two related clusters with two T. cruzi putative transporters, encoded by the genes TcCLB.504085.60 and TcCLB.511277.400. These two genes, together with their closest related gene, TcCLB.506509.40, were selected for complementation assays in Lec8 CHO mutant cells, which are deficient in UDP-Gal transport.
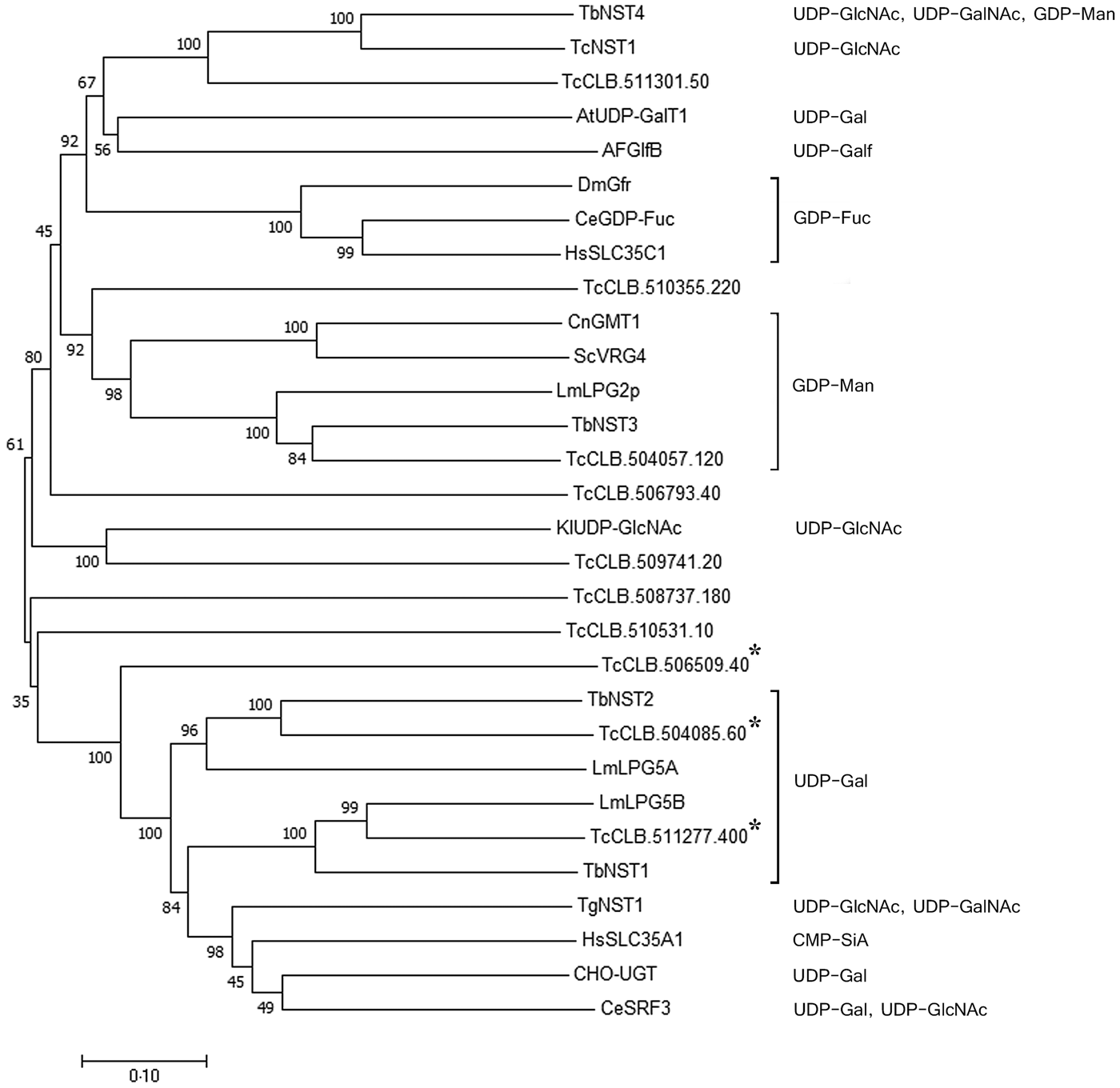
Fig. 1. Phylogenetic tree of functionally characterized NSTs and the putative T. cruzi NST family. Experimentally characterized substrates are indicated. The three T. cruzi genes selected as putative UDP-Gal transporters are indicated by asterisks. Sequences were aligned using Clustalx2.1, and the tree was constructed with Mega7.0.25. Statistical method: Neighbor-Joining. Model: p-distance. Number of bootstraps replications: 1000. Transporters are identified by substrate specificity and species. At, Arabidopsis thaliana; Ce, C. elegans; Cn, Cryptococcus neoformans; Dm, D. melanogaster; Hs, Homo sapiens; Kl, Kluyveromyces lactis; Lm, L. major; Sc, Saccharomyces cerevisiae; Tb, T. brucei; Tc, T. cruzi.
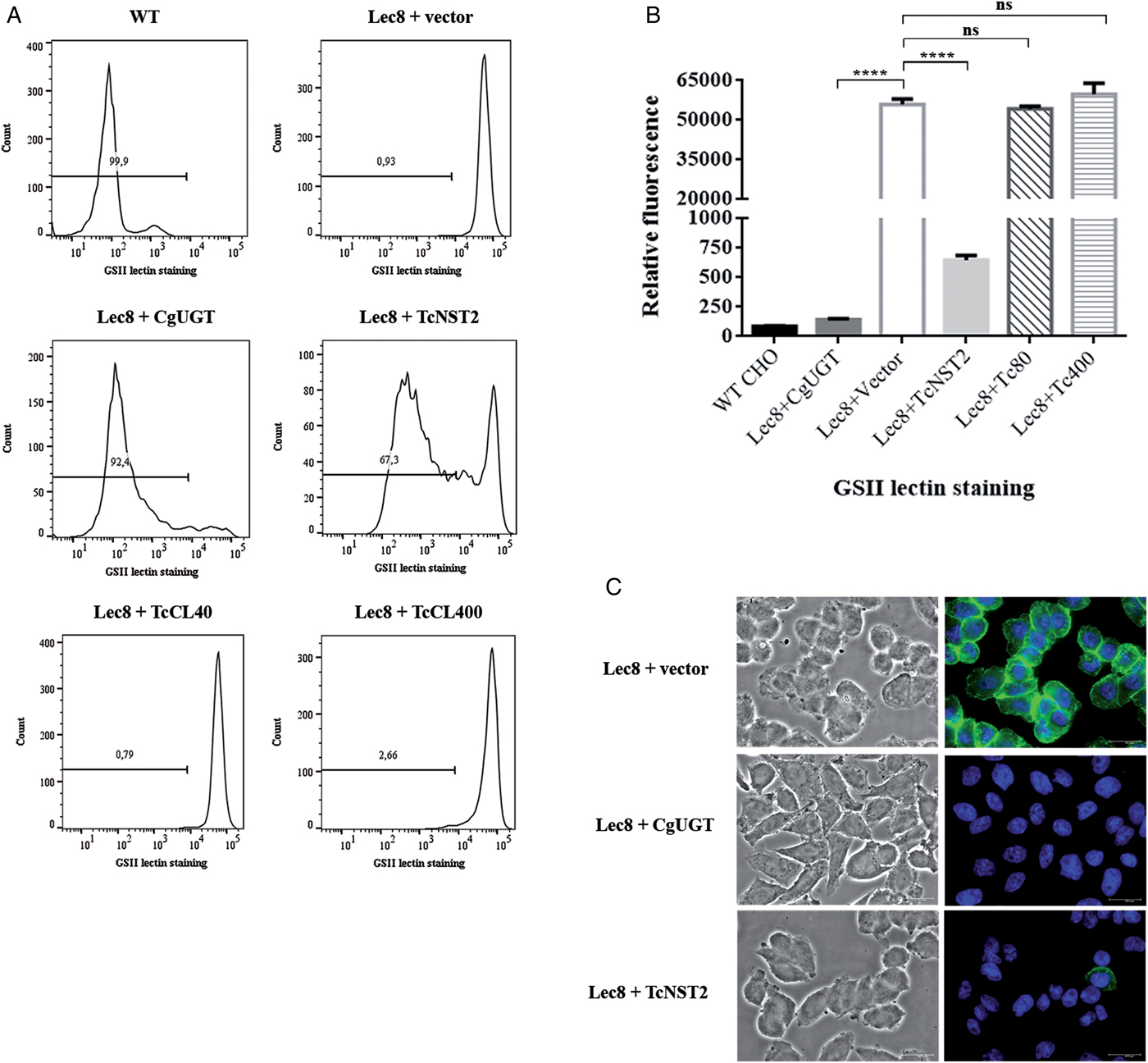
In wild-type CHO cells, galactose is attached to GlcNAc residues. In the mutant CHO cells, which are deficient in UDP-Gal transport (Deutscher and Hirschberg, Reference Deutscher and Hirschberg1986), GlcNAc residues are therefore more exposed due to a decreased galactosylation of proteins and lipids. In our assay, we tested the ability of the cells to bind the GS-II lectin from Griffonia simplicifolia, which specifically recognizes GlcNAc residues. Mutant cells bind more strongly to the lectin than the wild-type cells, as shown by flow cytometry analysis using GS-II conjugated to Alexa Fluor 488 (Fig. 2A).

Fig. 2. Identification of a UDP-Gal transporter (TcNST2) of T. cruzi by complementation of UDP-Gal transport-deficient Lec8 CHO cells. Mutant Lec8 cells were stably transfected with the T. cruzi genes TcCLB.504085.60 (TcNST2), TcCLB.506509.40 (TcCL40), and TcCLB.511277.400 (TcCl400), the endogenous CHO UDP-Gal transporter (CgUGT) or the empty vector (pcDNA3.1 (-)). (A) Representative histograms showing lectin binding of CHO cells labeled with Alexa Fluor 488-conjugated GS-II lectin and separated by flow cytometry. (B) Median fluorescence intensities of a representative experiment in triplicate. For TcNST2 and CgUGT, only cells in which fluorescence was shifted to wild-type levels were considered for plotting the graphs. Error bars represent standard deviations. ****, P < 0.0001; one-way ANOVA, Dunnett's multiple comparisons test. (C) Phase contrast images of stably transfected Lec8 cells fixed and incubated with GSII-Alexa Fluor 488 (green) and Hoechst 33258 DNA dye (blue).
The three selected T. cruzi candidate genes were stably expressed in Lec8 cells, and the binding of lectin was then analyzed by flow cytometry. Only the transporter encoded by the TcCLB.504085.60 gene (TcNST2) was able to rescue the wild-type phenotype (Fig. 2A). A decrease in GSII lectin binding to wild-type levels occurred in approximately 65% of cells. This partial restoration of the wild-type phenotype has also been observed in the heterologous expression of NSTs in yeast (Caffaro et al., Reference Caffaro, Hirschberg and Berninsone2006, Reference Caffaro, Luhn, Bakker, Vestweber, Samuelson, Berninsone and Hirschberg2008). Expression of the wild-type CHO UDP-Gal transporter in the same vector resulted in a shift of approximately 90% cells to wild-type levels of lectin binding. The other two T. cruzi genes or the empty vector had no effect on lectin binding. The median fluorescence intensity values of a representative experiment are presented in Fig. 2B (only cells in which fluorescence was shifted to wild-type levels were considered for plotting the graphs). Furthermore, the binding of lectin to TcNST2-transfected Lec8 cells was analyzed by fluorescence microscopy (Fig. 2C). Most of the cells transfected with TcNST2 showed no apparent binding to GS-II, as was observed in mutant cells expressing the endogenous CHO UDP-Gal transporter. As expected, mutant Lec8 cells transfected with the TcCLB.511277.400 and TcCLB.506509.40 genes (data not shown) or the empty vector were strongly bound to GS-II and showed a strong fluorescence signal.
The TcCLB.504085.60 gene and its allele, TcCLB.507089.40, are both composed of 1239 nucleotides and code for a 412-amino acid protein, displaying a hydropathy profile typical of the NST family (data not shown). Based on an epitope-insertion approach (Eckhardt et al., Reference Eckhardt, Gotza and Gerardy-Schahn1999), NSTs were predicted to have 10 transmembrane domains (TM), which was confirmed by the crystal structure of the S. cerevisiae GDP-Man transporter Vrg4 (Parker and Newstead, Reference Parker and Newstead2017). In order to have insights into the overall structure of TcNST2, a 3D model was generated using the I-TASSER server (Yang et al., Reference Yang, Yan, Roy, Xu, Poisson and Zhang2015). Superposition of the TcNST2 model with the crystal structure of Vrg4 and that of Vrg4 in complex with GDP-Man (PDB accession numbers 5OGE and 5OGK for the apo and GDP-Man bound structures, respectively) resulted in a TM-score of 0.6 (values above 0.5 indicating folding conservation) and 10% sequence identity, based on the structural alignment.
T. cruzi TcNST2 predicted topology is similar to that of Vrg4 and other members of the metabolite transporter superfamily, displaying a clear correspondence of the 10 TMs α-helices, and with the N- and C-termini facing the cytoplasmic side of the Golgi complex (Fig. 3A). However, the T. cruzi NST2 model reveals a long cytoplasmic insertion of approximately 80 residues between α-helices TM2 and TM3 (Fig. 3A). The occurrence of an insertion at this position has been previously predicted for the L. major UDP-Gal transporters LPG5A and LPG5B by transmembrane prediction algorithms (Capul et al., Reference Capul, Barron, Dobson, Turco and Beverley2007). Interestingly, the T. brucei UDP-Gal transporter TbNST2 also displays a large loop between TM domains 2 and 3. In order to further investigate these unusual large loops, which are not present in other NST members, and due to the fact that transmembrane prediction algorithms are not completely reliable (Reddy et al., Reference Reddy, Cho, Ling, Reddy, Shlykov and Saier2014), 3D models were generated for LPG5A and TbNST2 (data not shown). Based on their predicted structures, the insertions between TM2 and TM3 for TcNST2, LPG5A and TbNST2 are of approximately 80, 50 and 65 residues, respectively. According to their supposed cytoplasmic localization, these loops are rich in charged and polar residues (>50%) and have a negative grand average of hydropathy (GRAVY) value, typical of hydrophilic polypeptides (Kyte and Doolittle, Reference Kyte and Doolittle1982). Functional characterization of these loops my reveal relevant data concerning structure and function of NSTs in trypanosomatids.
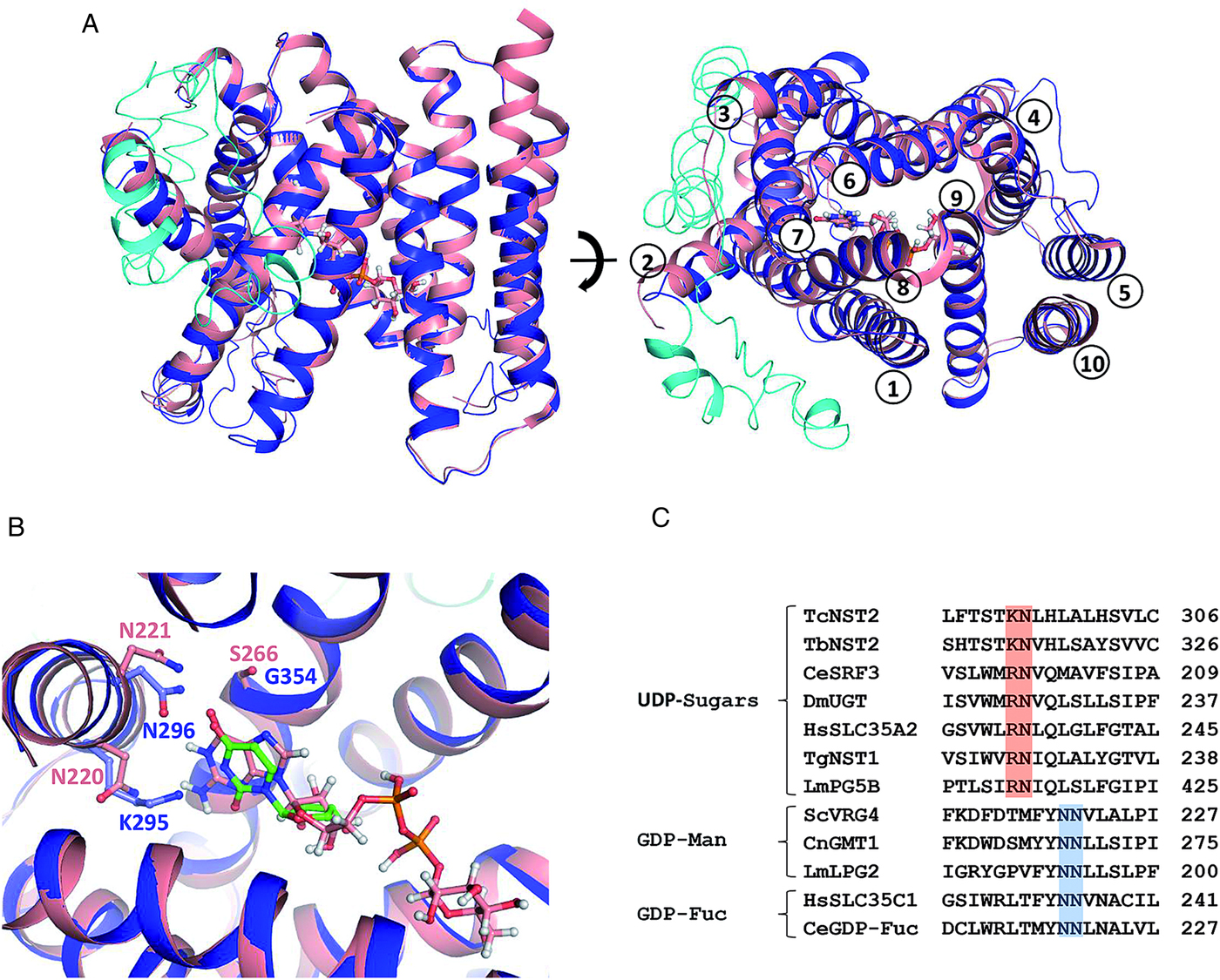
Fig. 3. Structural superposition of T. cruzi TcNST2 3D model (blue) and the crystal structure of S. cerevisiae Vrg4 (salmon) in complex with GDP-mannose (PDB 5OGK). (A) Overall view. TcNST2 insertion between helices TM2 and TM3 is shown in cyan. GDP-mannose, which was soaked into the Vrg4 crystals (Parker and Newstead, Reference Parker and Newstead2017), is represented in sticks. On the left panel the cytosol side is at the top of the figure. On the right panel the helices are numbered according S. cerevisiae Vrg4 topology. (B) Detail of the nucleotide sugar binding site. Residues involved in the interaction with the base moiety in ScVrg4 and corresponding residues in TcNST2 are shown in sticks and labeled. For comparison purposes an uridine (green) was manually superimposed to the GDP-mannose guanosine. (C) Conserved asparagine residues (N220 and N221) (blue), related to nucleotide recognition (Parker and Newstead, Reference Parker and Newstead2017), in GDP-sugar transporters. An asparagine (N220) is replaced by a conserved basic residue (K or R) (red), among UDP-sugar transporters of different organisms. TbNST2 (T. brucei, UDP-Gal/UDP-GlcNAc); CeSRF3 (C. elegans, UDP-Gal/UDP-GlcNAc); DmUGT (D. melanogaster, UDP-Gal/UDP-GalNAc); HsSLC35A2 (H. sapiens, UDP-Gal); TgNST1 (Toxoplasma gondii, UDP-GlcNAc/UDP-GalNAc); LmLPG5B (L. major, UDP-Gal); ScVrg4 (S. cerevisiae, GDP-Man); CnGMT1 (C. neoformans, GDP-Man); LmLPG2 (L. major, GDP-Man/GDP-Fuc/GDP-arabinose); HsSLC35A1 (H. sapiens, GDP-Fuc); CeGDP-Fuc (C. elegans, GDP-Fuc).
Another interesting feature revealed by the TcNST2 model is related to the nucleotide binding pocket. In S. cerevisiae Vrg4, N220 and N221 (from helix TM7) and S266 (from helix TM8) participate in the interaction with the guanine moiety (Parker and Newstead, Reference Parker and Newstead2017). In TcNST2 model an asparagine is conserved at the corresponding position of Vrg4 N221. However, a lysine residue (K295) occupies the position of Vgr4 N220 (Fig. 3B). Alignment of TcNST2 and other UDP-sugar transporters revealed conservation of a basic residue at this position (either lysine or arginine) (Fig. 3C). S266 is substituted with a glycine in TcNST2 which is also conserved in UDP-sugar transporters (data not shown).
TcNST2 subcellular localization and gene expression
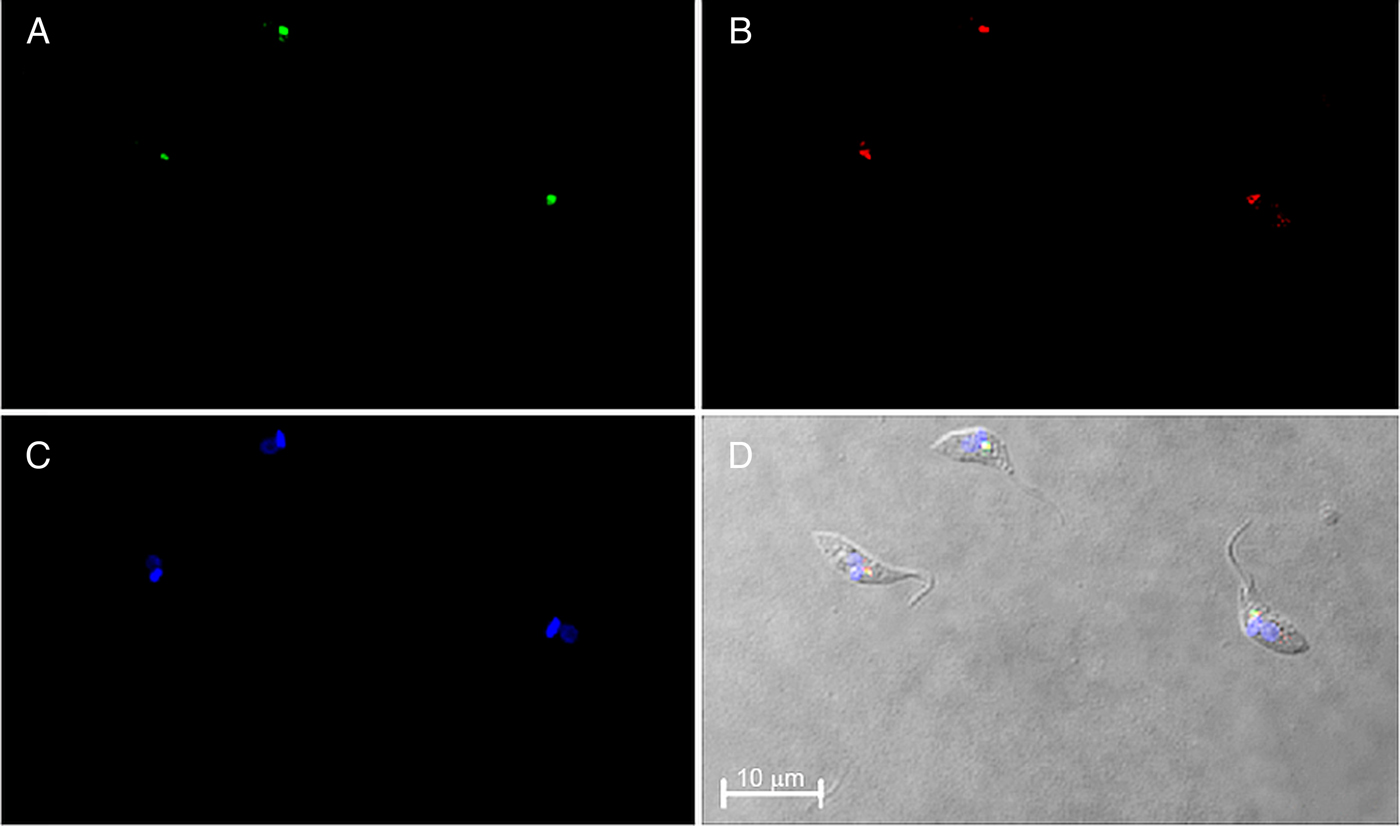
The subcellular localization of TcNST2 was investigated by fluorescence microscopy using a chimeric protein with GFP at the C-terminus of the transporter. A monoclonal antibody against GFP was used for the microscopic analysis due to weak signals obtained with the intrinsic fluorescence of GFP after fixation of the parasites. In neomycin-resistant epimastigotes of T. cruzi stably expressing the chimeric transporter, a clear signal at the anterior region of the parasite was observed (Fig. 4A). It is in close contact with the kinetoplast, as would be expected for a Golgi-resident protein (Fig. 4D). The subcellular localization of TcNST2 was confirmed by colocalization with TcHIP (Fig. 4B and D), a putative T. cruzi zDHHC palmitoyl transferase, whose Golgi localization was determined with the T. cruzi Golgi marker TcRab7 (Batista et al., Reference Batista, Kalb, Moreira, Batista, Eger and Soares2013).
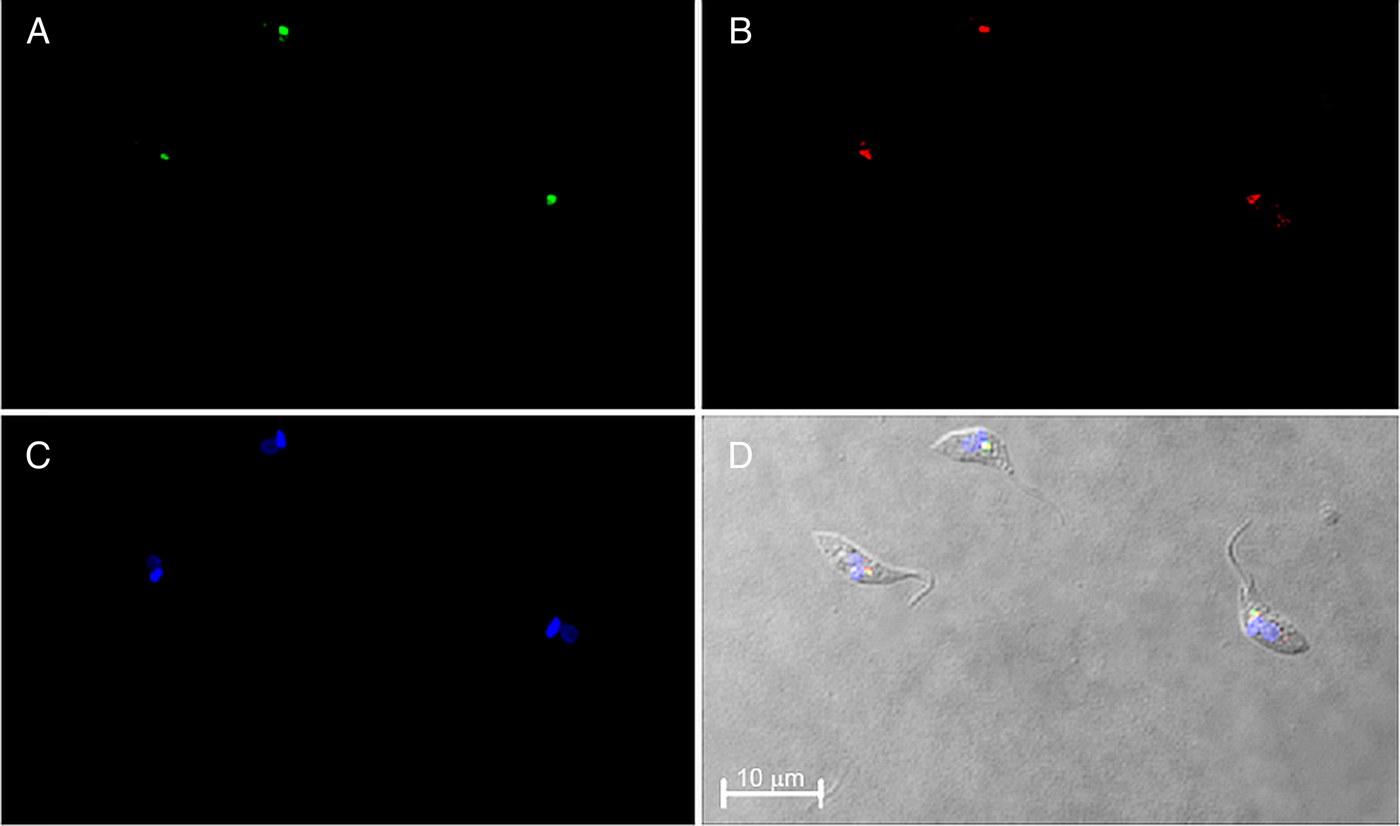
Fig. 4. Immunolocalization of TcNST2 to the Golgi apparatus by confocal laser microscopy. T. cruzi epimastigotes were transfected with a plasmid encoding a chimeric protein with GFP at the C-terminus of the transporter (TcNST2-GFP). Exponentially grown epimastigotes expressing the fusion protein were incubated with a monoclonal antibody anti-GFP (A) or an antiserum against T. cruzi TcHIP, a Golgi marker (B). Kinetoplasts and nuclei were stained with DAPI (C). Colocalization is shown in the DIC image merged with A, B and C (D).
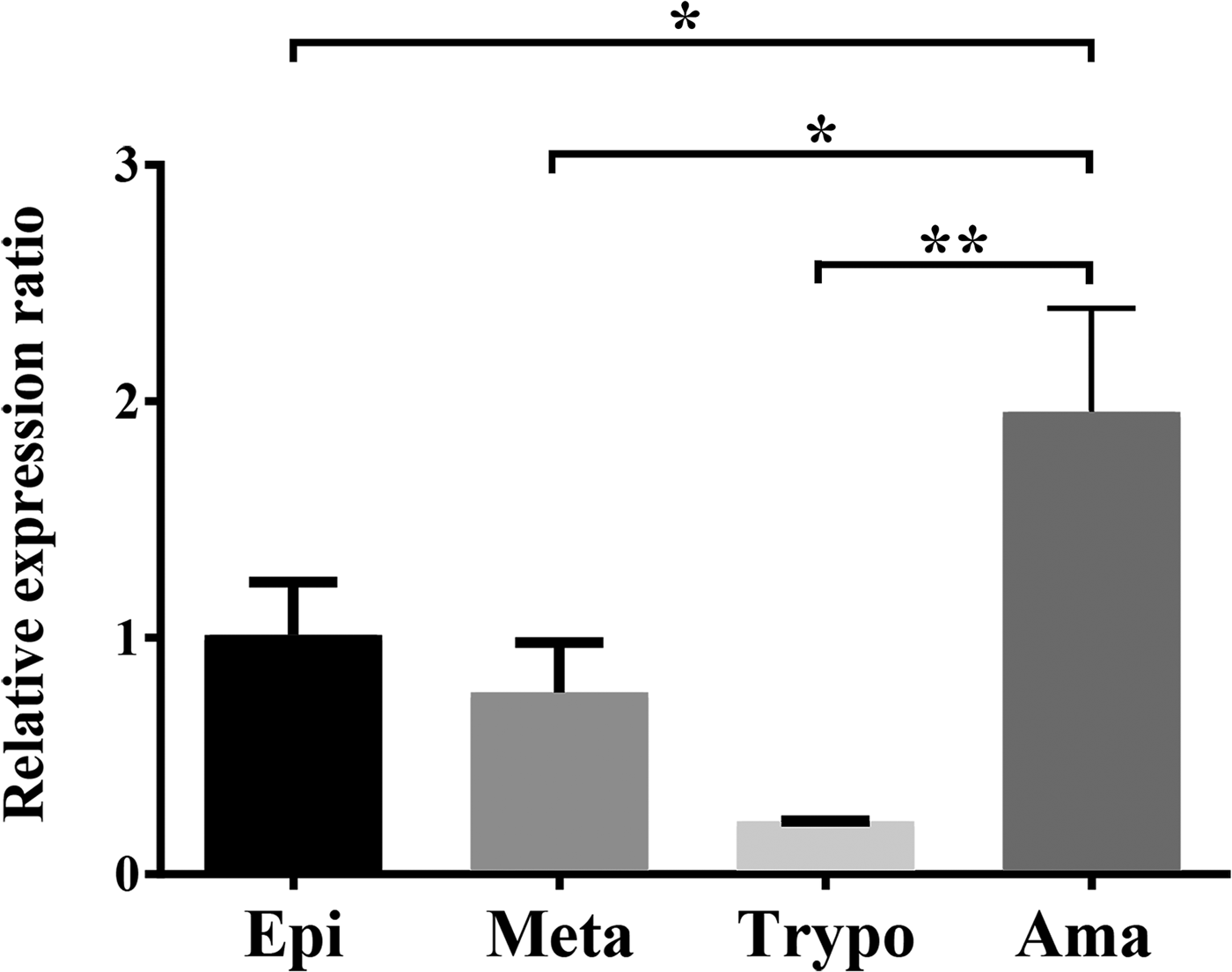
Expression of the TcNST2 gene was analyzed by quantitative PCR along the life cycle of T. cruzi. TcNST2 mRNA is detected in all four major life-cycle forms of T. cruzi, displaying a statistically higher expression level in axenic amastigotes (Fig. 5).
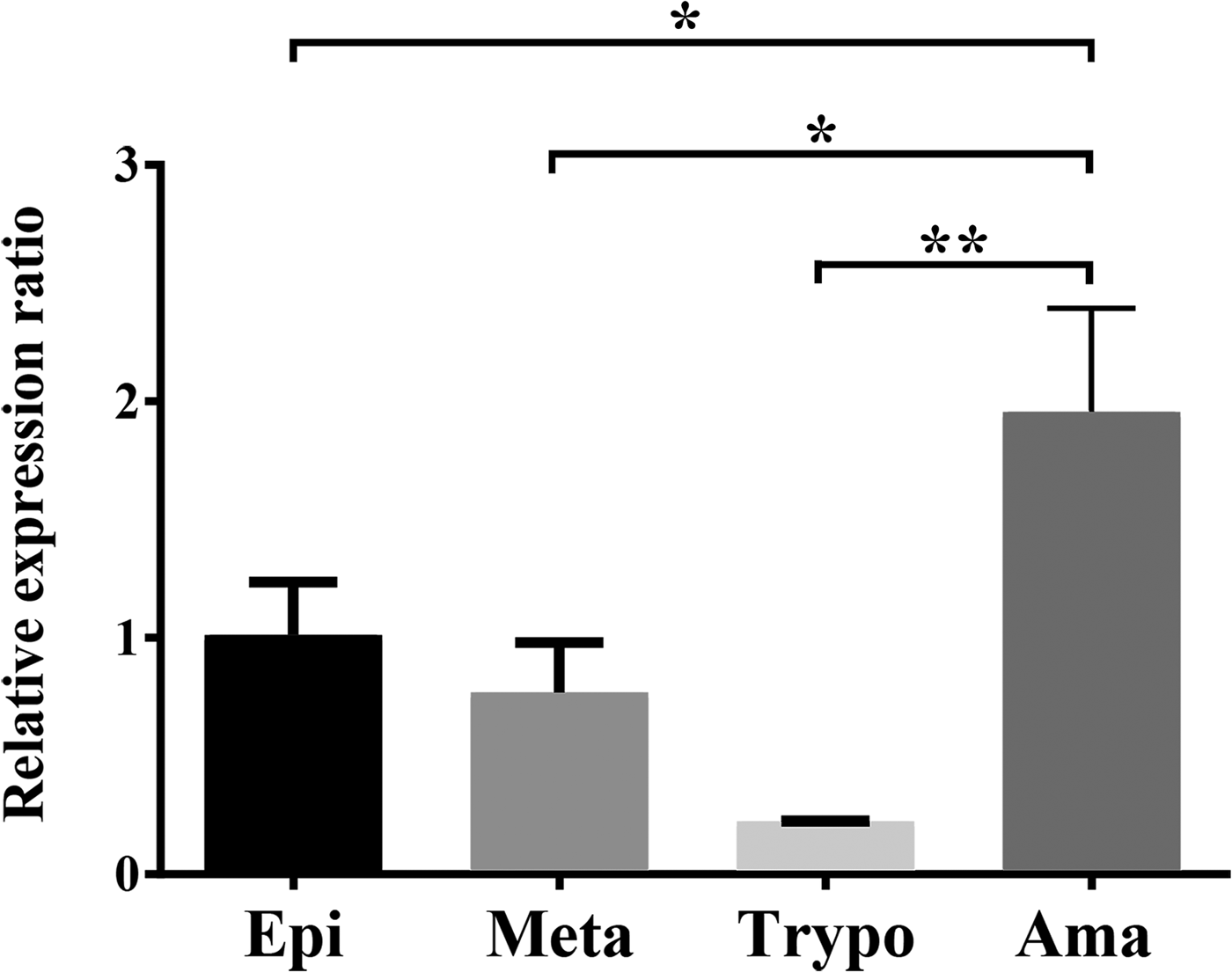
Fig. 5. TcNST2 expression in all life-cycle forms of T. cruzi. qPCR analysis of TcNST2 mRNA levels in epimastigotes (Epi), metacyclic trypomastigotes (Meta), cell culture-derived trypomastigotes (Trypo) and axenic amastigotes (Ama). Expression of the 60S ribosomal protein L9 and histone 2B was used as an internal control as described in Materials and Methods. Error bars represent standard deviations based on the number of biological replicates. *, 0.01<P < 0.05; **, 0.001<P < 0.01; one-way ANOVA, Dunnett's multiple comparisons test.
Discussion
To identify UDP-Gal transporters in T. cruzi, three candidate genes closely related to UDP-Gal transporters from other trypanosomatids were selected and tested in a complementation assay in CHO mutant cells. This mutant cell line has been used for the identification of UDP-Gal transporters in a variety of organisms, including Caenorhabditis elegans (Caffaro et al., Reference Caffaro, Luhn, Bakker, Vestweber, Samuelson, Berninsone and Hirschberg2008), Drosophila melanogaster (Segawa et al., Reference Segawa, Kawakita and Ishida2002) and L. major (Capul et al., Reference Capul, Barron, Dobson, Turco and Beverley2007). Only one transporter, TcNST2, restored the wild-type phenotype of the mutant cells (Fig. 2). It is important to note, however, that T. cruzi may have other UDP-Gal transporters since we only tested three putative NSTs in our screening assay.
In our previous work (Baptista et al., Reference Baptista, Rodrigues, Morking, Klinke, Zardo, Soares, de Aguiar, Goldenberg and Ramos2015), TcNST2 was tested as a putative UDP-GlcNAc transporter but failed to complement Kluyveromyces lactis mutant cells defective in the intracellular transport of this substrate. Therefore, TcNST2 probably transports UDP-Gal in T. cruzi, but the complementation test suggests that it is not a UDP-GlcNAc transporter. Interestingly, the T. brucei transporter TbNST2, which is the closest experimentally characterized transporter to TcNST2 (59% in sequence similarity), transports both UDP-Gal and UDP-GlcNAc (Liu et al., Reference Liu, Xu, Caradonna, Kruzel, Burleigh, Bangs and Hirschberg2013). This difference in substrate specificity between two closely related transporters is in accordance with the established difficulty in determining substrate specificity of NSTs based on sequence or phylogenetic analysis.
The predicted 3D model of TcNST2 (Fig. 3A and B) is remarkably similar to that of the GDP-Man transporter Vrg4, revealing an overall structural conservation in the NST family. Interestingly, we noted that an asparagine residue that is well-conserved in GDP-sugar transporters and related to nucleotide recognition (Parker and Newstead, Reference Parker and Newstead2017), is replaced with a conserved basic residue (lysine or arginine) in transporters for UDP-sugars (Fig. 3C). We hypothesize whether the substitution of asparagine by a longer and basic side chain may be related to the recognition mechanism of the uracil base by TcNST2. Structural comparison with the Vrg4 nucleotide binding pocket suggests that TcNST2 K295 side chain could be in a favorable position to interact with the uracil carbonyl group. The importance of this conserved basic residue for UDP-sugar transporters remains to be experimentally confirmed.
Nucleotide sugars are mostly synthesized in the cytosol with the exception of CMP-sialic acid, whose last enzymatic step occurs in the nucleus (Caffaro and Hirschberg, Reference Caffaro and Hirschberg2006). In trypanosomatids, however, nucleotide sugars are supposed to be synthesized in glycosomes based on the glycosomal localization of enzymes involved in their biosynthesis (Stokes et al., Reference Stokes, Guther, Turnock, Prescott, Martin, Alphey and Ferguson2008). If this is indeed the case, there must be another step of transport through the glycosomal membrane. The nature and identification of these transporters, however, have yet to be discovered. TcNST2 is structurally similar to other NSTs and was unequivocally localized to the Golgi apparatus by colocalization with TcHIP (Fig. 4). This intracellular localization suggests that TcNST2 is, at least in part, responsible for providing the pool of UDP-Gal inside the Golgi apparatus. In T. cruzi mucins, the O-linked glycans are attached to the polypeptide backbone via a GlcNAc residue, whose addition to threonine residues is catalyzed by a Golgi-localized O-GlcNAc transferase (Previato et al., Reference Previato, Sola-Penna, Agrellos, Jones, Oeltmann, Travassos and Mendonca-Previato1998; Heise et al., Reference Heise, Singh, van der Wel, Sassi, Johnson, Feasley, Koeller, Previato, Mendonca-Previato and West2009; Koeller et al., Reference Koeller, van der Wel, Feasley, Abreu, da Rocha, Montalvão, Fampa, Dos Reis, Atella, Souto-Padrón, West and Heise2014). Galactose residues inside the Golgi lumen are then added to GlcNAc by specific galactosyl transferases. These enzymes have not yet been studied in T. cruzi, but their activities have already been described in T. brucei (Pingel and Duszenko, Reference Pingel and Duszenko1992; Pingel et al., Reference Pingel, Rheinweiler, Kolb and Duszenko1999; Ramasamy and Field, Reference Ramasamy and Field2012) and Leishmania (Sizova et al., Reference Sizova, Ross, Ivanova, Borodkin, Ferguson and Nikolaev2011).
Expression of the TcNST2 gene throughout the parasite's life cycle was investigated in total RNA samples by qPCR. Our results suggest that the transporter is expressed in all life-cycle forms and is specifically upregulated in axenic amastigotes (Fig. 5). The amastigote surface is decorated with galactose residues, being intensely labeled with purified Chagasic anti-α-Gal antibodies (Souto-Padron et al., Reference Souto-Padron, Almeida, de Souza and Travassos1994). The upregulated mRNA levels detected in axenic amastigotes may reflect expression of the transporter for the proper galactosylation of amastigote glycoconjugates. The downregulation of TcNST2 mRNA levels observed in trypomastigotes is less clear since bloodstream trypomastigote mucins are known to contain a high number of α-Galp residues, which are the main target of anti-α-galactosyl antibodies present in the blood of infected patients (Almeida et al., Reference Almeida, Milani, Gorin and Travassos1991, Reference Almeida, Ferguson, Schenkman and Travassos1994; Gazzinelli et al., Reference Gazzinelli, Pereira, Romanha, Gazzinelli and Brener1991).
In trypanosomatids, however, gene expression is regulated mainly at the posttranscriptional level, with mRNA stability (Alves and Goldenberg, Reference Alves and Goldenberg2016) and translational efficiency (Smircich et al., Reference Smircich, Eastman, Bispo, Duhagon, Guerra-Slompo, Garat, Goldenberg, Munroe, Dallagiovanna, Holetz and Sotelo-Silveira2015) being important control points. As a consequence mRNA levels may not represent the actual protein content in the cell. In metacyclic trypomastigotes, transcriptional activity (Ferreira et al., Reference Ferreira, Dossin, Ramos, Freymüller and Schenkman2008) and translation (Smircich et al., Reference Smircich, Eastman, Bispo, Duhagon, Guerra-Slompo, Garat, Goldenberg, Munroe, Dallagiovanna, Holetz and Sotelo-Silveira2015) are decreased in comparison with epimastigotes. Actually, Ferreira et al. (Reference Ferreira, Dossin, Ramos, Freymüller and Schenkman2008) showed that the transcriptional rate remained unchanged during the intermediary steps of metacyclogenesis being reduced only when metacyclics were formed. It is possible that, due to the reduced rate of transcription and translation, proteins required in metacyclics are synthetized previously in epimastigotes, or during the differentiating process into metacyclic trypomastigotes. Transcriptional activity is also decreased in cell-derived trypomastigotes when compared to amastigotes (Elias et al., Reference Elias, Marques-Porto, Freymüller and Schenkman2001). We speculate that TcNST2 may be synthesized in amastigotes, in which mRNA levels are upregulated, and that the transporter remains active during the differentiation of amastigotes to trypomastigotes to supply the Golgi pool of UDP-Gal for the incorporation of galactose residues to amastigote and trypomastigotes glycoconjugates. In particular, in a study of the role of 3′UTR sequences of stage-specific regulated genes of T. cruzi, a trans-sialidase gene was shown to be upregulated at the protein level in trypomastigotes with a concomitant strong decrease in mRNA levels (Parker and Newstead, Reference Parker and Newstead2017).
On the other hand, it is also possible that the lower TcNST2 mRNA levels detected in trypomastigotes may indeed reflect the transporter expression in this life-cycle form. In this case, the transport of UDP-Gal through the Golgi membrane could be carried out by another as yet not identified UDP-Gal transporter. We have previously identified a family of eleven putative nucleotide sugar transporters in T. cruzi (Baptista et al., Reference Baptista, Rodrigues, Morking, Klinke, Zardo, Soares, de Aguiar, Goldenberg and Ramos2015) but, as stated before, we have only tested three putative NSTs in our screening assay for UDP-Gal transporters.
Finally, we must also consider that axenic amastigotes, obtained from the in vitro differentiation of cell-derived trypomastigotes by low pH (Tomlinson et al., Reference Tomlinson, Vandekerckhove, Frevert and Nussenzweig1995), may not reflect accurately the surface composition and the process of glycoconjugate synthesis of in vivo intracellular amastigotes. Even though intracellular and axenic amastigotes are morphologically similar and share the stage-specific surface antigen Ssp-4 (Tomlinson et al., Reference Tomlinson, Vandekerckhove, Frevert and Nussenzweig1995), we cannot rule out that our qPCR data may not correspond to the TcNST2 mRNA levels in intracellular amastigotes.
With respect to the insect-dwelling life-cycle forms of T. cruzi, our qPCR data suggest a more constitutive level of expression, in accordance with a study comparing the transcriptome (using polyadenylated RNA) and the translatome (ribosome profiling) of epimastigotes and metacyclic trypomastigotes (Smircich et al., Reference Smircich, Eastman, Bispo, Duhagon, Guerra-Slompo, Garat, Goldenberg, Munroe, Dallagiovanna, Holetz and Sotelo-Silveira2015). In conclusion, based on its specificity, subcellular localization and expression pattern, it is tempting to speculate that TcNST2 may play an important role in the synthesis of mucins and other galactose-containing glycoconjugates in T. cruzi.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182019000738.
Author ORCIDs
Augusto S. P. Ramos, 0000-0003-3925-2507.
Acknowledgements
We are grateful to Dr Maurilio J. Soares for helpful discussions and Dr Daniela P. Pavoni for technical assistance with the analysis of qPCR data (both located at the Instituto Carlos Chagas – Fiocruz PR). In addition, we would thank the Fiocruz Network of Technology Platforms for the use of core facilities.
Financial support
This work was supported through grants from the Fundação Oswaldo Cruz – Fiocruz; the Conselho Nacional de Desenvolvimento e Tecnológico – CNPq (grant number 482755/2009-1); the Fundação Araucária – FA (grant number 335/2012); the Programa de Pesquisa para o SUS – PPSUS (Decit/SCTIE/MS, FA and SESA-PR, grant number 1004/2013); the Programa Estratégico de Apoio à Pesquisa em Saúde – PAPES VI (Fiocruz) and the Programa de Apoio a Núcleos de Excelência – PRONEX (Acordo CNPq/FA).
Conflict of interest
None.
Ethical standards
Polyclonal antibodies were produced in mice according to international guidelines for the ethical use of animals.