INTRODUCTION
Parasites exhibit a wide range of life-history strategies that contribute to different dispersal abilities, host specialization, transmission modes, life-cycle complexity and population structure. Population genetic approaches can be used to understand the ecology and evolution of single species and, by recognizing the impact of host population genetic structure on that of the parasite, comparative studies of interacting species are becoming more common (e.g. McCoy et al. Reference McCoy, Boulinier and Tirard2005; Gómez-Díaz et al. Reference Gómez-Díaz, González-Solís, Peinado and Page2007, Reference Gómez-Díaz, Morris-Pocock, Gonzales-Solis and McCoy2012; Whiteman et al. Reference Whiteman, Kimball and Parker2007; Bruyndonckx et al. Reference Bruyndonckx, Henry, Christe and Kerth2009; Jones and Britten Reference Jones and Britten2010; Stefka et al. Reference Stefka, Hoeck, Keller and Smith2011).
The findings from population genetic analyses of hosts and parasites are as variable as the nature of the interactions themselves. Congruence between host and parasite population genetic structure (or lack of structure) depends on relative rates of host and parasite dispersal, host specificity of the parasite, host and parasite geographical distribution as well as a myriad of ecological factors that can influence hosts and parasites (Dybdahl and Lively, Reference Dybdahl and Lively1996; Johnson et al. Reference Johnson, Williams, Drown, Adams and Clayton2002; McCoy et al. Reference McCoy, Boulinier, Tirard and Michalakis2003; Weckstein, Reference Weckstein2004). Parasites are often cited as having higher evolutionary potential compared to their hosts due to shorter generation times and, in some cases, faster mutation rates (Page et al. Reference Page, Lee, Becher, Griffiths and Clayton1998). In an obligate, host-specific parasite, this could lead to greater population differentiation in the parasite relative to the host. This pattern has been shown across a wide range of host–parasite interactions, from a host plant and fungal pathogen (Delmotte et al. Reference Delmotte, Bucheli and Shykoff1999), seabird and tick ectoparasite (McCoy et al. Reference McCoy, Boulinier and Tirard2005), raptor and lice and fly ectoparasites (Whiteman et al. Reference Whiteman, Kimball and Parker2007) to butterflies and specialist parasitoids (Anton et al. Reference Anton, Zeisset, Musche, Durka, Boomsma and Settele2007). However, there are also a number of examples showing the opposite pattern: parasites that exhibit less population differentiation than their hosts, e.g. a freshwater snail and Schistosoma parasite (Davies et al. Reference Davies, Webster, Krüger, Munatsi, Ndamba and Woolhouse1999), stinging nettle and its parasitic plant (Mutikainen and Koskela, Reference Mutikainen and Koskela2002), two shearwater seabirds and their louse and flea ectoparasites (Gómez-Díaz et al. Reference Gómez-Díaz, González-Solís, Peinado and Page2007) and prairie dogs and their flea ectoparasites (Jones and Britten, Reference Jones and Britten2010). Untangling the factors acting on both hosts and parasites that contribute to these disparate patterns is important for understanding the context of coevolutionary interactions.
Seabirds provide a good system to investigate population differentiation in hosts and parasites. Seabirds are often very philopatric (Friesen et al. Reference Friesen, Burg and McCoy2007), which can contribute to strong population differentiation despite high potential vagility (Levin and Parker, Reference Levin and Parker2012). Many seabirds are large-bodied, and harbour high numbers of diverse groups of parasites (Hughes and Page, Reference Hughes and Page2007). We investigated the population genetic structure of two seabird ectoparasites (Hippooscidae), Olfersia spinifera and O. aenescens, relative to the differing degree of population genetic structure found in their respective hosts (Levin and Parker, Reference Levin and Parker2012), great frigatebirds (Fregata minor) and Nazca boobies (Sula granti), in the Galapagos Islands, Ecuador. Great frigatebirds and Nazca boobies are common throughout the Galapagos archipelago, breeding in medium to large colonies, often alongside closely related seabirds such as the magnificent frigatebird (Fregata magnificens) and blue-footed (Sula nebouxii) and red-footed (Sula sula) boobies.
Hippoboscid flies are highly specialized obligate parasites of birds and mammals that typically spend all of their adult life on the host. Most of the species that infect birds have functional wings and are capable of flying between individual hosts (Harbison et al. Reference Harbison, Jacobsen and Clayton2009; Harbison and Clayton, Reference Harbison and Clayton2011); however, little is known about their dispersal tendencies. Hippoboscid flies feed on host blood several times a day (Coatney, Reference Coatney1931) and can have negative effects on host health, such as anaemia (Jones, Reference Jones1985) and slow chick development (Bishopp, Reference Bishopp1929). In addition to impacting hosts by feeding on blood, hippoboscids are also vectors for Haemoproteus parasites (Levin et al. Reference Levin, Valkiunas, Santiago-Alarcon, Cruz, Iezhova, O'Brien, Dearborn, Schreiber, Fleischer, Ricklefs and Parker2011) and trypanosomes (Baker, Reference Baker1967). Flies belonging to the genus Olfersia are typically found infecting frigatebirds and boobies, and in our sampling of over 300 hippoboscid flies from five species of seabird, we have never found O. spinifera on a booby host or O. aenescens on a frigatebird.
There is convincing evidence that O. spinifera is the main vector of a haemosporidian parasite, Haemoproteus iwa, that infects frigatebirds throughout their geographical range (Levin et al. Reference Levin, Valkiunas, Santiago-Alarcon, Cruz, Iezhova, O'Brien, Dearborn, Schreiber, Fleischer, Ricklefs and Parker2011). Interestingly, we have found no genetic differentiation in the haemosporidian parasite across this range (Levin et al. Reference Levin, Valkiunas, Santiago-Alarcon, Cruz, Iezhova, O'Brien, Dearborn, Schreiber, Fleischer, Ricklefs and Parker2011) despite strong genetic differentiation between Galapagos frigatebirds (F. magnificens and F. minor) and their non-Galapagos conspecifics (Hailer et al. Reference Hailer, Schreiber, Miller, Levin, Parker, Chesser and Fleischer2011, Hailer, unpublished data). It is possible that the broad distribution of this one H. iwa lineage could be facilitated by movement of infected O. spinifera. Therefore, we predicted less population genetic structure in O. spinifera than in the bird host, F. minor. We used S. granti and O. aenescens as a comparison, because multilocus and mitochondrial data show strong differentiation between populations of S. granti at the small geographical scale within the Galapagos Islands, while F. minor shows weak to no differentiation (Levin and Parker, Reference Levin and Parker2012). If hippoboscid flies are moving between individuals at roosting or non-breeding sites, we expect to find more gene flow in both fly species relative to gene flow in the bird hosts, regardless of the strength of host population genetic structure.
MATERIALS AND METHODS
Sampling
Fregata minor and S. granti and their fly ectoparasites from six different islands (Darwin, Española, Genovesa, North Seymour, San Cristobal and Wolf) in the Galapagos Archipelago were sampled during June and July of 2007, 2008, 2010 and 2011. Española and North Seymour were visited twice, once during 2007 and again in 2010. The investigated seabird colonies were all mixed-species colonies with different membership. Fregata minor, S. granti and the red-footed booby, S. sula, are found nesting in sympatry on Genovesa, Darwin and Wolf. The island of Española is a breeding ground for F. minor (although very few nests were found in 2010), S. granti and the blue-footed booby (S. nebouxii). Both great frigatebirds (F. minor) and magnificent frigatebirds (F. magnificens) are found breeding on North Seymour, along with S. nebouxii. San Cristobal is the one breeding colony in Galapagos where one can regularly find all three booby species breeding sympatrically. Fregata minor were seen roosting on San Cristobal in 2011, but no evidence of nesting was noted. Although breeding is not synchronous throughout the archipelago, there were typically sufficient numbers of breeding F. minor and S. granti to sample (sample sizes can be found in Table 1). Seabirds were captured by hand and a small blood sample was taken from the brachial vein. Blood was preserved in lysis buffer at ambient temperature in the field and later stored at 4 °C in the laboratory. Birds were systematically searched for flies and, if present, at least 1 was collected and stored in 95% ethanol. We included a small number (n = 31) of flies sampled from non-focal, but sympatric hosts (O. aenescens from S. sula (n = 9, 3 islands) and S. nebouxii (n = 14, 2 islands), O. spinifera from F. magnificens (n = 8, one island)). Once in the laboratory, flies were kept at −20 °C until DNA extraction. Sampling was done in accordance with the University of Missouri – St. Louis IACUC guidelines.
Table 1. Sample sizes, number of haplotypes, haplotype diversity (h) and nucleotide diversity (π) for two seabird species, great frigatebird (Fregata minor) and Nazca booby (Sula granti) and their respective hippoboscid fly ectoparasites (Olfersia spinifera and O. aenescens)

DNA extraction and mitochondrial DNA amplification
DNA extraction and amplification of mtDNA (Cytochrome Oxidase I (COI), Cytochrome b (Cyt b), and NADH dehydrogenase 2 (ND2)) for seabird hosts has been described by Levin and Parker (Reference Levin and Parker2012) and seabird mtDNA sequences can be found in GenBank (Accession numbers: JX569150-JX569187).
We always analysed flies collected from separate host individuals. Olfersia spinifera sampled from F. minor on Española and Genovesa were so numerous that we included more flies in our analyses than birds from these islands, but never more than one fly from any individual bird. Thoraxes of hippoboscid flies were separated from the heads and abdomens. A Qiagen DNEasy Blood and Tissue DNA extraction kit (Qiagen, USA, Germantown MD) was used to individually extract the DNA from each fly thorax. The standard protocol was followed, but DNA was eluted in half as much buffer due to the assumed low concentrations of any parasite or host DNA. Undiluted DNA was used in PCR reactions. COI was amplified using LCO1490 and HCO2198 (Folmer et al. Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994) following the reaction conditions described in Whiteman et al. (Reference Whiteman, Sanchez, Merkel, Klompen and Parker2006) except for an annealing temperature of 46 °C rather than 40 °C. A region of mitochondrial 12S ribosomal DNA (12S) was amplified using the primer pair 12SAI and 12SBI (Simon et al. Reference Simon, Frati, Beckenback, Crespl, Liu and Flook1994) using the reaction conditions found in Whiteman et al. (Reference Whiteman, Sanchez, Merkel, Klompen and Parker2006). The primer pair L11122 and H11823 was used to amplify a portion of Cyt b following the protocol described by Page et al. (Reference Page, Lee, Becher, Griffiths and Clayton1998). Purification of the PCR product and subsequent sequencing was performed as described for the bird hosts. Fly mtDNA sequences can be found in GenBank (Accession numbers KC700559-KC700601).
Population genetic analyses
DNA sequences from flies were assembled and edited in Seqman 4.0 (DNASTAR, USA) and aligned by ClustlW implemented in BioEdit v7.0.5.3 (Hall, Reference Hall1999). All three mitochondrial gene regions were aligned separately, cropped, concatenated and analysed together for both hosts and parasites. Population equilibrium and selective neutrality were assessed using a Tajima's D-test (Tajima, Reference Tajima1989) in DNASP v.5.10.01 (Librado and Rozas, Reference Librado and Rozas2009). Minimum spanning haplotype networks were calculated using ARLEQUIN v3.5.1.2 (Excoffier et al. Reference Excoffier, Laval and Schneider2005), drawn using HapStar (Teacher and Griffiths, Reference Teacher and Griffiths2011) and coloured for clarity in Inscape v.0.48.2. Haplotype and nucleotide diversities were calculated in DNASP. Traditional F-statistics (Wright, Reference Wright1951) were used to assess variation within and between populations. Analysis of molecular variance (AMOVA, Excoffier et al. Reference Excoffier, Smouse and Quattro1992) was used to partition components of genetic variation among and within island populations. The number of migrants per generation (N m) was estimated from F ST values using Wright's formula (Wright, Reference Wright1951) and used to compare relative amounts of movement between the two bird hosts, the two fly parasites and between the respective bird–parasite pairs. If some level of population genetic differentiation was found, we tested for isolation by distance using Slatkin's linearized F ST (F ST/(1−F ST)) in the program IBD (Bohonak, Reference Bohonak2002).
RESULTS
F. minor and O. spinifera population genetic structure
Analysis of mitochrondrial DNA from the seabird hosts was completed as part of a multi-locus, comparative population genetic study (Levin and Parker, Reference Levin and Parker2012) and summarized here to make the comparison with the structure of the ectoparasites. A total of 1954 bp of mitochondrial DNA (after editing and cropping to equal length) were amplified for F. minor (Cyt b: 766 bp, ND2: 489 bp, COI: 699 bp) and 1608 bp were amplified for O. spinifera (Cyt b: 630 bp, 12S: 362 bp, COI: 616 bp). There was no indication of non-neutrality in F. minor sequence data (Tajima's D = −1·64, P>0·05) but O. spinifera sequences showed a significant departure from neutrality as determined by the Tajima's D test (D = −2·49, P<0·01). Fourteen variable sites were recovered from the F. minor sequence, seven of which were parsimony informative sites. In comparison, 27 variable sites were found in O. spinifera, only seven of which were parsimony informative sites. Sample sizes (total and per island), number of haplotypes, haplotype diversity (h) and nucleotide diversity (π) can be found in Table 1. For F. minor, the lowest haplotype diversity, 0·257, was found in the birds sampled from Darwin, and the highest was found in the North Seymour sample (0·783) (Levin and Parker, Reference Levin and Parker2012). For the frigatebird fly, O. spinifera, the lowest haplotype diversity was recovered from Wolf; however, we only captured two flies from this island. The island with the most diverse O. spinifera haplotypes was Genovesa (0·649). Haplotype networks for F. minor and O. spinifera can be found in Fig. 1.

Fig. 1. Haplotype networks for Galapagos great frigatebirds (Fregata minor) (A) and their obligate hippoboscid fly ectoparasite, Olfersia spinifera (B), constructed from mitochondrial DNA. Circles are proportional to the number of individuals that share that haplotype and colours correspond to different islands. Black = Darwin, blue = Wolf, green = Genovesa, yellow = North Seymour, red = Española. Frigatebird haplotype network is from Levin and Parker (Reference Levin and Parker2012).
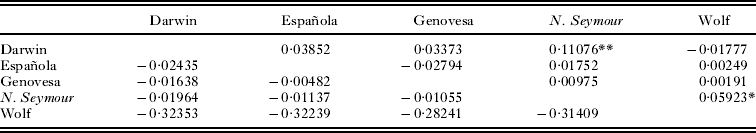
An analysis of molecular variance showed very weak population genetic structure in F. minor with only 2·29% of the variance partitioned among island populations and a global ϕST of 0·023 (Levin and Parker, Reference Levin and Parker2012). The AMOVA run on the O. spinifera dataset showed no support for any subdivision of genetic diversity (P = 0·971). Pair-wise F ST values for F. minor and O. spinifera can be found in Table 2. Two pair-wise comparisons (North Seymour – Darwin, North Seymour – Wolf) were significant for F. minor (Levin and Parker, Reference Levin and Parker2012). No pair-wise comparisons between any O. spinifera populations sampled indicated significant differentiation (Table 2). The estimated number of F. minor migrants per generation (N m) ranged from 4·01 between North Seymour and Darwin to an infinite number between Española and Genovesa and Darwin and Wolf. Olfersia spinifera show complete panmixia within Galapagos. There was no support for a pattern of isolation by distance between island populations of F. minor (Mantel tests, genetic distance vs geographical distance: z = 85·1, r = 0·34. P= 0·13) (Levin and Parker, Reference Levin and Parker2012).
Table 2. FST values from mtDNA for great frigatebirds (Fregata minor) above the diagonal and Olfersia spinifera ectoparasitic flies below the diagonal
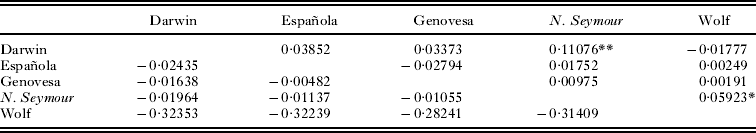
** P < 0·01; *P < 0·05.
Sula granti and O. aenescens population genetic structure
Slightly longer COI sequences were obtained for S. granti (799 bp) giving us a total amount of 2145 bp (Cyt b: 780 bp, ND2: 566) (Levin and Parker, Reference Levin and Parker2012). In total, 1671 base pairs of mitochondrial DNA were used for analyses of O. aenescens (Cyt b: 678, 12S: 361. COI: 632). Sula granti and O. aenescens sequence data showed no departure from neutrality (Tajima's D, S. granti: D = −1·00, P>0·05; O. aenescens: D = 1·75, P>0·05). Sample sizes (total and per island), number of haplotypes, haplotype diversity (h) and nucleotide diversity (π) can be found in Table 1. Very few flies were captured from S. granti; this species, like related Galapagos sulids, has fewer ectoparasites than Galapagos frigatebirds (Rivera et al. unpublished data). Overall, haplotype diversity in S. granti ranged from 0·644 on Española to 0·933 on Wolf (Levin and Parker, Reference Levin and Parker2012) (Table 3). The average haplotype diversity of O. aenescens was 0·830; however, that calculation is based on a small sample including only the three islands that had more than one haplotype sampled. Haplotype networks for S. granti and O. aenescens can be found in Fig. 2.

Fig. 2. Haplotype network for Galapagos Nazca boobies (Sula granti) (A) and their obligate hippoboscid fly ectoparasite, Olfersia aenescens (B), constructed from mitochondrial DNA. Circles are proportional to the number of individuals that share that haplotype and colours correspond to different islands. Black = Darwin, blue = Wolf, green = Genovesa, purple = San Cristobal, red = Española. Nazca booby haplotype network is from Levin and Parker (Reference Levin and Parker2012).
Table 3. FST values from mtDNA for Nazca boobies (Sula granti) above the diagonal and Olfersia aenescens ectoparasitic flies below the diagonal

** P < 0·01; *P < 0·05.
Analyses of molecular variance revealed significant genetic differentiation in S. granti but not O. aenescens; the among-population component was a good predictor of genetic partitioning in S. granti (P = 0·00098), explaining 13·49% of the variance (Levin and Parker, Reference Levin and Parker2012), while no differentiation was detected in O. aenescens (P = 0·808). Four of the 10 pair-wise FSTs (Darwin vs remaining 4 islands) were significant in the case of S. granti (Levin and Parker, Reference Levin and Parker2012), while no significant pair-wise comparisons were found for O. aenescens. The relative number of S. granti migrants per generation (N m) ranged from 1·39 in the case of migrants between Española and Darwin to an infinite number between Española and San Cristobal. Olfersia aenescens showed patterns of unrestricted gene flow across all population pairs, with the lowest N m estimate of 72·5 between Española and San Cristobal. This must be interpreted with caution due to small sample sizes. The genetic structure of S. granti populations did have some signature of isolation by distance, driven largely by the significant differentiation between Darwin, a peripheral island, and all other populations (Mantel test, genetic distance vs geographical distance: z = 491·84, r = 0·38, P = 0·02) (Levin and Parker, Reference Levin and Parker2012).
Flies from alternative hosts
A small number of flies were sampled from alternative hosts (O. spinifera from F. magnificens; O. aenescens from S. nebouxii and S. sula). In order to explore the possibility that these other hosts could be contributing to the population genetic patterns described above, we sequenced the same section of COI for these flies (O. aenescens: 632 bp; O. spinifera: 616 bp). Haplotypes and nucleotide diversities are reported in Table 1, although we do not have adequate samples sizes to calculate these statistics by island. Only one COI haplotype was found in O. spinifera collected from F. magnificens and it matched the most common COI haplotype from flies collected from F. minor. Six haplotypes were recovered from each sample of O. aenescens collected from the alternative booby hosts, three of which were shared between flies from both blue-footed and red-footed boobies. When compared with O. aenescens COI haplotypes from S. granti, three of nine haplotypes are shared among all three host species, one haplotype is shared among S. granti and S. sula, three haplotypes are unique to O. aenescens from S. nebouxii, and two haplotypes are unique to O. aenescens from S. sula.
DISCUSSION
Host movement has been shown to be a key determinant of parasite gene flow (Gómez-Díaz et al. Reference Gómez-Díaz, González-Solís, Peinado and Page2007, Reference Gómez-Díaz, Morris-Pocock, Gonzales-Solis and McCoy2012; McCoy et al. Reference McCoy, Beis, Barbosa, Cuervo, Fraser, González-Solís, Jourdain, Poisbleau, Quillfeldt, Léger and Dietrich2012). However, host movement is often assessed indirectly via population genetic studies that only reveal effective dispersal events, in which movement is followed by reproduction. By simultaneously applying these same indirect genetic assessments of gene flow to closely associated parasites, we increase our ability to detect host movement that is not necessarily tied to successful reproduction. Here we show that two obligate fly ectoparasite species have higher levels of gene flow than their respective host species, despite marked differences in the genetic structures of the host populations.
Relatively more genetic diversity was partitioned among island populations in the birds than in the flies. This pattern is evident in the haplotype networks . Interestingly, both the star-like structure of the O. spinifera network and the significant Tajima's D statistic indicate a recent, rapid expansion of this population. There are a number of possible explanations for this. It is possible that the population of frigatebirds colonizing the Galapagos were free of O. spinifera and acquired the parasite more recently; however, we have rarely handled a frigatebird that does not have at least one fly parasite. We have no reason to believe that non-Galapagos F. minor are less parasitized; their large bodies and high survival coupled with their non-diving behaviour makes them good hosts for ectoparasites (e.g. Felso and Rozsa, Reference Felso and Rozsa2006). Alternatively, recent expansion could be due to population bottlenecks caused by El Niño Southern Oscillation (ENSO) events that dramatically affect the climatic conditions in the Galapagos Islands (Valle et al. Reference Valle, Cruz, Cruz, Merlen and Coulter1987). Little is known about hippoboscid breeding biology, but it is possible that the increased precipitation associated with ENSO events could affect flies in their pupal stage, the only life stage that is off the host. If there is low survival of pupae and adult flies do not live until the next breeding season (related hippoboscid flies are estimated to live approximately 80–100 days; Nelson et al. Reference Nelson, Keirans, Bell and Clifford1975), this could contribute to a population bottleneck.
It is difficult to imagine that hippoboscid flies are able to disperse between islands without ‘hitchhiking’ on a bird host. We do know that, despite having restricted host breadth, O. spinifera are frequently moving between F. minor hosts on a local (within-island colony) scale (Levin and Parker, unpublished data). It is possible that the larger-scale fly movements indicated by these genetic data are facilitated by bird hosts other than the main ones we analysed here. Olfersia spinifera also parasitize magnificent frigatebirds (F. magnificens), which are found breeding on some islands in Galapagos and O. aenescens are reported from other Sulid species such as the blue-footed booby (S. nebouxii) and the red-footed booby (S. sula), both of which breed in Galapagos. Frigatebirds and booby species are often found nesting in mixed seabird colonies in Galapagos, but we have never found O. aenescens on frigatebirds or O. spinifera on booby species. Based only on cyt b sequence divergence, these two fly species differ by 8·5%. Ectoparasite dispersal via alternative hosts has been suggested in the black-tailed prairie dogs–flea (Oropsylla hirsuta) system where a similar pattern of higher ectoparasite gene flow relative to host gene flow was found (Jones and Britten, Reference Jones and Britten2010).
It is possible that S. granti's congeners, S. sula or S. nebouxii, could be moving O. aenescens around the archipelago. At the sites we sampled, there was at least one other species of Sulid breeding (Española: S. granti and S. nebouxii; Genovesa, Darwin, Wolf: S. granti and S. sula, San Cristobal: S. granti, S. nebouxii and S. sula). No genetic differentiation was found among S. nebouxii populations, based on a comparison of samples from three island colonies (Taylor et al. Reference Taylor, Maclagan, Anderson and Friesen2011). A comparison of three colonies of S. sula indicated significant differentiation between one pair of the islands (Darwin and Genovesa) (Baiao and Parker, unpublished data). We sequenced a fragment of COI for a small number of O. aenescens from S. sula and S. nebouxii to explore this possibility of movement by alternative hosts and to gain insight into whether different host races of O. aenescens exist as a result of specialization on the different Sulidae species. We recovered nine COI haplotypes from O. aenescens on the three booby species and the three most common haplotypes were shared among O. aenescens sampled from all three booby species, indicating that the pattern we report in O. aenescens on S. granti may be a result of movement via alternative hosts. However, we did find five haplotypes each unique to one S. sula host (two cases) or S. nebouxii host (three cases), indicating that there may be some degree of differentiation or specialization of flies on these hosts. We used the same approach with O. spinifera on F. magnificens, which breeds sympatrically with F. minor in one of our sampling locations (North Seymour). All eight O. spinifera collected from F. magnificens had the most common COI haplotype, shared by the majority of O. spinifera sampled from F. minor. Therefore, we cannot rule out that the population genetic patterns found in O. spinifera sampled from F. minor are affected by movements of this alternative host. Ideally, a comparison of multiple parasites from these hosts would be used to further tease apart the effects of host movement on parasite population genetic structure.
An alternative explanation of higher gene flow in both fly species compared with their bird hosts involves non-breeding movements of birds, including movement of juveniles, prospecting by young and breeding birds and movement by adult birds whose breeding attempt has failed (Gómez-Díaz et al. Reference Gómez-Díaz, Morris-Pocock, Gonzales-Solis and McCoy2012). Frigatebirds are not sexually mature until at least 5 years of age (Valle et al. 2006) and we do not know the extent of their movements prior to breeding. Even if they are philopatric to their natal site as has been suggested (Metz and Schreiber, Reference Metz, Schreiber, Poole and Gill2002; Dearborn et al. Reference Dearborn, Anders, Schreiber, Adams and Mueller2003), movement of juveniles prior to breeding age could facilitate ectoparasite dispersal. Frequent shorter, inter-island and long-distance movements of F. minor are reported both in the breeding season and during the non-breeding season (Dearborn et al. Reference Dearborn, Anders, Schreiber, Adams and Mueller2003). Friesen et al. (Reference Friesen, Burg and McCoy2007) found that the extent of population genetic structure in seabirds can be explained in part by non-breeding distributions. Philopatry to non-breeding areas appears to reduce or prevent gene flow between seabird populations (Friesen et al. Reference Friesen, Burg and McCoy2007). There is evidence from radio-telemetry data on post-breeding movements that suggests frigatebirds are not always philopatric to non-breeding sites (Weimerskirch et al. Reference Weimerskirch, le Corre, Marsac, Barbraud, Tostain and Chastel2006). Long-distance dispersal events have been recorded rarely in S. granti, with most breeding and natal dispersal distances on the order of 100 m or less (Huyvaert and Anderson, Reference Huyvaert and Anderson2004). Only two of close to 7000 S. granti banded at Punta Cevallos, Española were recaptured at a different colony: one on the other side of Española 14 km away, and one approximately 200 km from the natal colony on Genovesa (Huyvaert and Anderson, Reference Huyvaert and Anderson2004). In both cases, the birds appeared to be visiting and not breeding and the bird located on Genovesa was seen at Punta Cevallos a week later (Huyvaert and Anderson, Reference Huyvaert and Anderson2004). Subadult S. granti are known to leave their natal colony post-fledging for up to 3 years until they begin to breed (Huyvaert and Anderson, Reference Huyvaert and Anderson2004). During this period, they may be moving O. aenescens around the archipelago.
Theory predicts that gene flow is an important force for introducing novel genetic variation into populations (Gandon et al. Reference Gandon, Capoweiz, Dubois, Michalakis and Olivieri1996) and it has been suggested that greater relative rates of dispersal in parasites compared with their hosts should increase parasite local adaptation (Gandon and Michalakis, Reference Gandon and Michalakis2002). Studies of black-legged kittiwakes (Rissa tridactyla) show that relative gene flow in hosts and parasites (in this case the tick, Ixodes uriae) are scale dependent (McCoy et al. Reference McCoy, Boulinier, Schjørring and Michalakis2002, Reference McCoy, Boulinier and Tirard2005). Tick gene flow was similar or higher than kittiwake gene flow at a regional scale, but more restricted at a larger scale (McCoy et al. Reference McCoy, Boulinier and Tirard2005). Similarly, in parasites with complex life cycles and multiple hosts, dispersal in the most motile definitive host has been shown as the determining factor in the emergence of population genetic structure of the parasite (Louhi et al. Reference Louhi, Karvonen, Rellstab and Jokela2010). The higher levels of gene flow we detected among O. spinifera and O. aenescens populations may promote local adaptation, as suggested by host–parasite simulation research (Gandon and Michalakis, Reference Gandon and Michalakis2002). However, Gandon and Michalakis (Reference Gandon and Michalakis2002) indicate that there is a point at which migration is too high and the potential for local adaptation decreases. Additionally, local adaptation has been shown to be strongly affected by both host and parasite population sizes, with large populations increasing the level of adaptation (Gandon and Michalakis, Reference Gandon and Michalakis2002). While there are undoubtedly more hippoboscid flies than bird hosts in our system, their relatively long generation time may decrease the opportunity for local adaptation. Finally, it is important to consider that most O. aenescens haplotypes are shared between all three host species of booby and we find only a few hints of host specialization based on O. aenescens haplotypes unique to either S. sula or S. nebouxii. If there is substantial movement of O. aenescens between host species, this could favour any traits that make O. aenescens a better generalist.
Extensive movement of hippoboscid flies could have implications for the spread of other parasites throughout the ranges of these seabird species. Some lice and mites have been shown to attach to hippoboscids and disperse phoretically (e.g. Whiteman et al. Reference Whiteman, Sanchez, Merkel, Klompen and Parker2006; Harbison et al. Reference Harbison, Jacobsen and Clayton2009). High gene flow among hippoboscid fly populations may have profound effects on the potential for geographical expansion by the Haemoproteus parasites they vector. We hypothesize that the gene flow in hippoboscid flies demonstrated here could explain the presence of the one ubiquitous lineage of haemosporidian parasite, H. iwa, in frigatebirds sampled throughout their range (Levin et al. Reference Levin, Valkiunas, Santiago-Alarcon, Cruz, Iezhova, O'Brien, Dearborn, Schreiber, Fleischer, Ricklefs and Parker2011), suggesting frequent contact between frigatebirds from different breeding colonies on a large geographical scale. Even though Galapagos frigatebird species are genetically isolated compared with their conspecifics (Hailer et al. Reference Hailer, Schreiber, Miller, Levin, Parker, Chesser and Fleischer2011; Hailer et al. unpublished data), their parasite populations might be connected through host movements not associated with breeding more frequently than we would expect given host population genetic patterns alone.
ACKNOWLEDGEMENTS
We would like to thank S. Harden, C. McKinley, M. Favazza, A. Carrion, M. Evans, P. Baiao, J. Merkel, J. Higashiguchi, J. Pogacnik, S. O'Brien, S. Deem and J. L. Rivera for their participation in the fieldwork associated with this research. We thank the Galapagos National Park for sampling permission and the Charles Darwin Foundation for logistical support, particularly S. Cisneros. This manuscript was improved by suggestions from and discussions with members the Parker Laboratory group and by thoughtful comments from two anonymous reviewers. This publication is contribution number 2063 of the Charles Darwin Foundation for the Galapagos Islands.
FINANCIAL SUPPORT
The work was supported by the Des Lee Collaborative Vision and by two grants from the Field Research for Conservation programme of the Saint Louis Zoo, as well as grants to I. L. from the Whitney R. Harris World Ecology Center, Sigma Xi, and the Frank M. Chapman Memorial Fund of the American Museum of Natural History. I. L. completed this work while supported by a Dissertation Fellowship from the University of Missouri – St. Louis.