INTRODUCTION
The genus Bartonella includes bacteria formerly designated as Grahamella or Rochalimaea (Brenner et al. Reference Brenner, O'Connor, Winkler and Steigerwalt1993; Birtles et al. Reference Birtles, Harrison, Saunders and Molyneux1995), and today consists of 38 named or candidate (proposed) species according to a comprehensive survey of published literature (PubMed and Web of Knowledge). These intracellular erythrocytic microparasites are fastidious, aerobic, Gram-negative small rods in the family Alphaproteobacteria, phylum Rhizobiales, and are most closely related to Brucella, Agrobacterium, and Rhizobium species. A variety of Bartonella species have been isolated from a diverse array of wildlife worldwide, such as deer, elk, rodents, shrews, rabbits, lions, cheetahs, foxes, coyotes, raccoons, and kangaroos (Kosoy, Reference Kosoy2010; Kaiser et al. Reference Kaiser, Riess, O'Rourke, Linke and Kempf2011). Species have also been described from humans (Minnick and Battisti, Reference Minnick and Battisti2009; Vayssier-Taussat et al. Reference Vayssier-Taussat, Le Rhun, Bonnet and Cott2009; Kaiser et al. Reference Kaiser, Riess, O'Rourke, Linke and Kempf2011), from domestic companion animals such as dogs and cats and from livestock (cattle and sheep) (Breitschwerdt et al. Reference Breitschwerdt, Maggi, Chomel and Lappin2010). Reservoir host infections with a co-adapted bartonellae are typified by the presence of a persistent, non-pathogenic bacteraemia which is thought to facilitate bacterial acquisition by arthropod vectors (Birtles, Reference Birtles2005). Co-adaptation of the bacteria and host, or mutual adaptation resulting in bacterial propagation in the host with minimal detriment, is suggested by this phenomenon, and by field and laboratory evidence for host specificity of some Bartonella species for particular hosts (Kosoy et al. Reference Kosoy, Regnery, Tzianabos, Marston, Jones, Green, Maupin, Olson and Childs1997, Reference Kosoy, Regnery, Kosaya and Childs1999, Reference Kosoy, Saito, Green, Marston, Jones and Childs2000). Despite the presence of this long-term bacteraemia most evidence suggests that infection of a mammalian host with its co-adapted bartonella results in little, if any, deleterious effect to that host (Fichet-Calvet et al. Reference Fichet-Calvet, Jomaa, Ben Ismail and Ashford2000; Bai et al. Reference Bai, Calisher, Kosoy, Root and Doty2011), and if disease occurs in naturally infected hosts it is at low incidence (Birtles, Reference Birtles2005).
Though bartonella bacteria identified from wildlife sources are increasingly being implicated as zoonotic disease agents (Chomel and Kasten, Reference Chomel and Kasten2010), their natural history is not well understood. Rodents in particular are reservoirs for numerous bartonella strains and species (Kosoy, Reference Kosoy2010). Longitudinal field studies of rodents provide insight into population infection incidence and prevalence, yet the dynamics of transmission in these systems remain obscure. For example, the minimum bacterial dose required to initiate an infection in a rodent host is unknown, as is the extent to which individual hosts might exhibit different responses following infection (Kosoy et al. Reference Kosoy, Mandel, Green, Marston and Childs2004a, Reference Kosoy, Mandel, Green, Marston, Jones and Childsb). Vector transmission of several Bartonella species has been demonstrated (Chomel et al. Reference Chomel, Kasten, Floyd-Hawkins, Chi, Yamamoto, Roberts-Wilson, Gurfield, Abbott, Pedersen and Koehler1996; Bown et al. Reference Bown, Bennet and Begon2004; Cotte et al. Reference Cotte, Bonnet, Le Rhun, Le Naour, Chauvin, Boulouis, Lecuelle, Lilin and Vayssier-Taussat2008; Reis et al. Reference Reis, Cote, Le Rhun, Lecuelle, Levin, Vayssier Taussat and Bonnet2011), but alternate transmission mechanisms have not been completely ruled out, and deserve consideration as they may be responsible for some proportion of transmission events in nature. Bartonellae have been isolated from cotton rat and deer mouse embryos and neonates which suggests a role for vertical transmission in the natural maintenance cycles of these strains (Kosoy et al. Reference Kosoy, Regnery, Kosaya, Jones, Marston and Childs1998). Rodents are also known to excrete pathogens such as Leptospira or Hantavirus spp. in their urine, which could also be a mechanism for environmental transmission of bartonella bacteria (Meerburg et al. Reference Meerburg, Singleton and Kijlstra2009).
No studies have been published describing experimental infection of laboratory mice with bartonellae isolated from Mus spp. in nature. To better understand bartonella infection dynamics in natural hosts it would be desirable to develop a model system that pairs a natural host with its co-adapted bartonella bacteria. As Mus musculus is the definitive laboratory animal model a system comprised of the laboratory mouse and a co-adapted bartonella would have great utility for research purposes. To date, the well-documented host specificity of bartonellae has been an impediment to developing such a model (Karem et al. Reference Karem, Paddock and Regnery2000; Kosoy et al. Reference Kosoy, Saito, Green, Marston, Jones and Childs2000).
With an overall goal of obtaining insight into the natural history characteristics of rodent-bartonellae systems, we designed a study to evaluate the in vivo infection characteristics of 2 Mus species B. tribocorum strains in laboratory mice. Since these strains were originally obtained from wild-caught Mus species (M. caroli and M. cervicolor), we thought it likely that the bacteria could successfully switch from one Mus species to another, i.e. to M. musculus. Previously, we attempted to infect laboratory mice with Bartonella strains obtained from Rattus spp. (Colton et al. Reference Colton, Zeidner and Kosoy2011). Although 1 of 4 strains in that study did infect the mice, the infection dynamics of the strain in the mice were not consistent with our knowledge of naturally acquired infections of rodents (Kosoy et al. Reference Kosoy, Mandel, Green, Marston and Childs2004a, Reference Kosoy, Mandel, Green, Marston, Jones and Childsb; Bai et al. Reference Bai, Calisher, Kosoy, Root and Doty2011; Colton et al. Reference Colton, Zeidner and Kosoy2011). Since host specificity of rodent bartonellae has been demonstrated at the genus level, but not at the species level (Kosoy et al. Reference Kosoy, Saito, Green, Marston, Jones and Childs2000), we thought it probable that laboratory mouse infection with these Mus spp. B. tribocorum strains would produce a model exhibiting characteristics of naturally acquired infection (Kosoy et al. Reference Kosoy, Mandel, Green, Marston and Childs2004a, Reference Kosoy, Mandel, Green, Marston, Jones and Childsb; Telfer et al. Reference Telfer, Clough, Birtles, Bennett, Carslake, Helyar and Begon2007b). Specific goals for the study were to observe (1) whether CD1 (ICR) mice (M. musculus) would be susceptible to infection with Mus spp. bartonella strains, (2) the time to manifestation of bacteraemia following inoculation of a range of bacterial doses, (3) the duration and magnitude of bacteraemia in infected mice and, (4) whether viable bacteria might be present in bacteraemic mouse urine. A model with these characteristics would be desirable for investigating the dynamics of persistent bacteraemia and comparing host specificity of different Bartonella species. If mice were susceptible to infection at low doses of the bacteria, this model would also be useful for evaluating vector transmission of bartonellae among rodent hosts.
MATERIALS AND METHODS
Bacteria
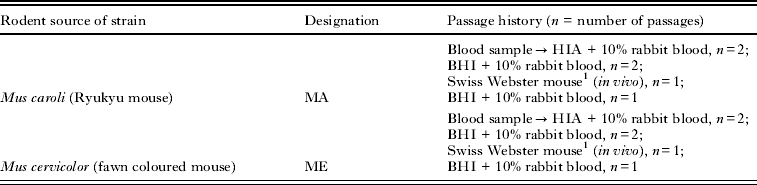
At the time of their original isolation from blood, the 2 strains used in this study were determined by genetic sequencing of a portion of the gltA gene to be most closely related to B. tribocorum. Passage history and other information for the 2 B. tribocorum strains used in the study is detailed in Table 1 and does not include growth of stock for this study. Although the 2 strains have nearly identical nucleotide sequence identities for their citrate synthase genes both were included in the study as they differed in host of origin. Bacteria for the study were grown on heart infusion agar plates (HIA) supplemented with 5% rabbit blood. Plates were inoculated to produce bacterial lawns and placed in a CO2 incubator at 35 °C for 5 days, at which time the bacteria were harvested in physiological saline. Stocks were then frozen at −80 °C until used for mouse inoculation. Bacterial stock concentrations were calculated by thawing aliquots of frozen stock, diluting them 10-fold, and inoculating them onto plates as described above to enumerate colony forming units (CFU).
Table 1. The Bartonella strains used in the study were originally isolated from Mus species in Thailand and have the same passage history

1 Mouse was infected with 106 cfu of bacteria, divided between intraperitoneal and subcutaneous inoculation sites.
Mice, experimental design, and blood collections
Thirty-six specific pathogen-free, 8-week-old CD1 female mice were obtained from the closed, outbred mouse colony at the Division of Vector-Borne Diseases (DVBD), Fort Collins, Colorado, USA. Mice were housed 6 to a cage and groups were segregated based on strain identity and dose for the study duration of 27 weeks. All work with the mice was approved by, and conducted under the supervision of DVBD's Institutional Animal Care and Use Committee (protocol 07-004), in accordance with United States Public Health Service standards for the humane care of laboratory animals.
Three groups of 6 mice were each subcutaneously inoculated with a different dose of bacteria: 10, 100, or 1000 cfu, for each of the 2 bacterial strains (n = 18 mice × 2 strains = 36). Bacterial doses were diluted in physiological saline to a volume of 300 μl. Mice were inoculated in the dorsal midline between the scapulae.
Blood was collected weekly from each mouse in the experiment for 27 weeks. Blood sample volume ranged from 80 to 125 μl depending on mouse body mass. Blood was collected by lancet puncture of the submandibular vessel plexus while the mouse was anaesthetized (ketamine-medetomidine cocktail [50–75 mg/kg + 1 mg/kg], administered intraperitoneally). The cheek of the mouse was first shaved and cleansed with chlorhexidine to create an aseptic site for blood collection, and the puncture site was alternated weekly. Following blood collections mice were administered the reversal agent atipamazole (5 mg/kg intraperitoneally). Blood was frozen at −80 °C until thawed for microbiological testing.
A high mouse mortality rate was observed during this study [6/36 (17%)], an unprecedented number in our experience of working with mice. All deaths occurred while mice were under anaesthesia. It was later learned that the ketamine used in our anaesthesia cocktail was recalled due to an unduly high number of adverse events including deaths related to its use (Teva Animal Health, 2009).
Urine collections
Urine was collected from mice weekly for 4 weeks, from weeks 3–6 of the study. Some, but not all mice were bacteraemic during this period. While mice were anaesthetized for blood collection, the bladder was gently squeezed and any urine expressed was captured in a sterile tube. Cystocentesis was not performed to preclude the possibility of lacerating blood vessels during urine collection, thus contaminating the sample with blood. Urines were frozen at −80 °C until thawed for microbiological plating.
Attempts were made to collect urine from all mice in the study but some mice voided during handling prior to anaesthesia induction. Collection volumes were extremely variable for those that had urine in the bladder and ranged from ∼20 to 70 μl per mouse. On a weekly basis urine was collected from 73–100% of mice.
Testing of samples to detect viable bartonella bacteria
Frozen whole blood was thawed and mixed 1:2 (50 μl + 50 μl) with a brain heart infusion (BHI) diluent containing 5% Fungizone™. The resultant 100 μl samples were inoculated onto HIA plates supplemented with 5% rabbit blood, and incubated for up to 2 weeks at 35 °C in a 5% CO2 atmosphere. Samples with bacterial colonies too numerous to count were subsequently diluted so that cfu could be counted and bacteraemia levels could be calculated. Colonies were confirmed as bartonella bacteria based on Gram stain and colony morphology (both rough and smooth) (Kosoy et al. Reference Kosoy, Regnery, Tzianabos, Marston, Jones, Green, Maupin, Olson and Childs1997). Mice whose blood samples grew 1 or more bacterial colonies were considered bacteraemic. The level of detection of bacteria by plating of samples was 20 cfu/ml of blood.
Urine samples were thawed and 50 μl of each sample was mixed with 50 μl of BHI diluent containing 8% Fungizone™. When the sample volume was less than 50 μl, all available urine was used for plating. The diluted samples were inoculated onto HIA plates supplemented with 5% rabbit blood, and held for up to 2 weeks at 35 °C in a 5% CO2 incubator.
RESULTS
Bacteraemia kinetics in mice
Mice were susceptible to infection with both Mus bartonella strains evaluated and displayed a dose-dependent response to both with ⩾60% of inoculated mice becoming bacteraemic (12/18 MA and 11/18 ME) (Table 2, Fig. 1A–E). Mice inoculated with the 1000 cfu dose of MA manifested bacteraemias as early as 1–2 weeks post-inoculation, whereas a temporal lag of at least 5–6 weeks occurred before any mouse manifested bacteraemia with ME regardless of dose (Fig. 1A–E, Table 2). Since some mice in the study died under anaesthesia, our bacteraemia data are incomplete. Mice that died apparently uninfected (no positive bacteraemia test prior to death) might have manifested late onset bacteraemias had they survived (n = 4/6; the other 2 mice were bacteraemic). For the 2 mice that died bacteraemic, their duration of bacteraemia is unknown. Three MA10, one MA100, one ME10, and one ME100 mouse died between weeks 4 and 15 of the study. The bacteraemia data that were collected for the mice that died are presented along with data from surviving mice (Table 2, Fig. 1A–E).

Fig. 1. (A) Bacteraemia kinetics for SW mice inoculated with strain MA (10 bacteria per mouse). Mice were inoculated at week 0, and did not manifest bacteraemia at our level of detection until 12 or more weeks post-inoculation. (B) Bacteraemia kinetics for SW mice inoculated with strain MA (100 bacteria per mouse). All mice inoculated at this dose that manifested a bacteraemia showed a temporal lag before testing positive. Mouse 2–5 was negative until week 19. (C) Bacteraemia kinetics for SW mice inoculated with strain MA (1000 bacteria per mouse). Mice inoculated with this dose displayed faster onset of bacteraemia (4/6 positive by week 3), late onset of bacteraemia (Mouse 3–4 and Mouse 3–6), and abacteraemic intervals (for example: Mouse 3–1, Mouse 3–2, and Mouse 3–4). (D) Bacteraemia kinetics for SW mice inoculated with strain ME (100 bacteria per mouse). As observed in mice inoculated with strain MA, mice in this ME-inoculated group displayed temporal lags in bacteraemia onset, and abacteraemic intervals. (E) Bacteraemia kinetics for SW mice inoculated with strain ME (1000 bacteria per mouse). Compared to mice in the MA group that were inoculated with 1000 bacteria per mouse, all mice in this group displayed temporal lags before onset of detectable bacteria. Abacteraemic intervals were also observed in mice in this group (for example Mouse 3–3).
Table 2. Summarized bacteraemia data for mice inoculated with low doses of 2 different mouse bartonella strains

1 Eight mice were still bacteraemic when the study was terminated: duration of bacteraemia data is therefore incomplete for some mice.
Bacteraemia duration in some individual mice was months long. We defined the duration as the time from first manifestation of bacteraemia until last bacteraemic observation or until the end of the study. This was regardless of whether bacteraemia was detected every single week between onset and termination. Some mice displayed abacteraemic intervals, or a pattern of relapsing bacteraemia (Fig. 1A–E; for example: Fig. 1A, Mouse 1–1). These intervals were characterized by detectable bacteraemia levels, followed by periods without detectable bacteraemia (level of detection was 20 cfu/ml of blood), after which bacteraemias were once again detected (Fig. 1A–E; for example: Fig. 1A, Mouse 1–1; Fig. 1B, Mouse 2–5). The profile for Mouse 1–1 is an example of a relapsing pattern of bacteraemia, with this mouse displaying 3 abacteraemic intervals. Bacteraemia began at week 15, then was not detected again until week 19, then was undetectable until week 22, and then undetectable again until week 24. After week 24 bacteraemia was not detected again, and the study ended at week 27. Abacteraemic intervals were often associated with an overall pattern of decreasing bacteraemia levels (Fig. 1A–E; for example: Fig. 1B, Mouse 2–6).
Inoculated mice in both groups showed variability in time to bacteraemia onset (Fig. 1A–E, Table 2). This was especially evident in the MA strain group at lower doses (10 or 100 cfu), where temporal lags in manifestation of detectable bacteraemia ranged from several weeks to months for mice within the same dose group (for example: Fig. 1A, Mouse 1–1; Fig. 1B, Mouse 2–5). Some mice were still bacteraemic when the study was terminated (8/30 surviving mice) (for example: Fig. 1D, Mouse 2–1 and 2–5).
Presence of bacteria in urine
During the 4-week collection period urine was collected from each mouse in the study at least 3 times. Urine was successfully collected from some of the mice all 4 times. No bartonella bacteria were detected in any of the mouse urines by microbiological plating.
DISCUSSION
This is the first report of experimental infection of laboratory mice with Mus spp. bartonella strains, a homologous host-bacteria system at the genus level. These B. tribocorum strains were easily able to cross-species host switch from their hosts of origin to the laboratory mouse. Persistent bacteraemias were observed in a high proportion of inoculated mice following exposures ranging from 10 to 1000 cfu. Infected laboratory mice manifested high-level, long-duration bacteraemias with abacteraemic intervals reproducing characteristics of infection reported for natural reservoirs of rodent bartonella strains (Kosoy et al. Reference Kosoy, Mandel, Green, Marston, Jones and Childs2004b; Birtles, Reference Birtles2005; Bai et al. Reference Bai, Calisher, Kosoy, Root and Doty2011).
Longitudinal field studies looking at populations of field voles (Microtus agrestis) in the UK, cotton rats (Sigmodon hispidus) in Georgia, USA, and deer mice (Peromyscus maniculatus) in Colorado, USA have documented bacteraemias from several months to a year duration in naturally infected rodents (Bai et al. Reference Bai, Calisher, Kosoy, Root and Doty2011; Birtles, Reference Birtles2005; Kosoy et al. Reference Kosoy, Mandel, Green, Marston and Childs2004a). Bacteraemia levels reported for naturally infected rodent hosts range from 106 cfu/ml for bartonella strains infecting Sigmodon hispidus (cotton rats) (Kosoy et al. Reference Kosoy, Mandel, Green, Marston and Childs2004a), to levels of 105 cfu/ml in various Rattus spp. (Castle et al. Reference Castle, Kosoy, Lerdthusnee, Phelan, Bai, Gage, Leepitakrat, Monkanna, Khlaimanee, Chandranoi, Jones and Coleman2004), to 104 cfu/ml in Neotoma albigula and N. micropus (woodrats) (Morway et al. Reference Morway, Kosoy, Eisen, Montenieri, Sheff, Reynolds and Powers2008) and Cynomys ludovicianus (black-tailed prairie dogs) (Bai et al. Reference Bai, Kosoy, Ray, Brinkerhoff and Collinge2008).
Mice in our study exhibited high, sustained levels of bacteraemia following low-dose exposures and represent the first immunocompetent mouse model to replicate the longevity of bartonella infection well beyond 11 weeks. Two other laboratory mouse models for persistent bacteraemia have been reported (Boulouis et al. Reference Boulouis, Barrat, Bermond, Bernex, Thibault, Heller, Fontaine, Piemont and Chomel2001; Koesling et al. Reference Koesling, Aebischer, Falch, Schulein and Dehio2001; Marignac et al. Reference Marignac, Barrat, Chomel, Vayssier-Taussat, Gandoin, Bouillin and Boulouis2010). The more extensively investigated of the two uses a strain of B. birtlesii originally isolated from Apodemus species (Boulouis et al. Reference Boulouis, Barrat, Bermond, Bernex, Thibault, Heller, Fontaine, Piemont and Chomel2001; Marignac et al. Reference Marignac, Barrat, Chomel, Vayssier-Taussat, Gandoin, Bouillin and Boulouis2010). Laboratory mice infected with this strain manifest bacteraemias up to 10 weeks in duration with levels of bacteria in the blood up to 105 cfu/ml. Immunocompetent mice inoculated intravenously with 109 cfu of B. grahamii developed bacteraemias up to 104 cfu/ml and 11 weeks duration (Koesling et al. Reference Koesling, Aebischer, Falch, Schulein and Dehio2001). Alterations in infection response relative to natural host infection kinetics, such as truncated or low-level bacteraemias even following high-dose bacterial exposures, are probably due to suboptimal levels of bacterial adaptation to an alternate host.
Temporal lags in bacteraemia onset associated with different dose exposures were commonly observed during this study. The dynamics of response seemed to be primarily influenced by the dose inoculated, where lower doses generally seem to elicit shorter duration bacteraemias of somewhat lower levels. Lags before bacteraemia onset following bacterial exposure may be a common characteristic of natural infections, and may complicate attempts to define the transmission dynamics of rodent bartonella strains among their hosts. There is evidence that bartonella bacteria are transmitted by fleas (Chomel et al. Reference Chomel, Kasten, Floyd-Hawkins, Chi, Yamamoto, Roberts-Wilson, Gurfield, Abbott, Pedersen and Koehler1996; Bown et al. Reference Bown, Bennet and Begon2004), yet analyses of data collected during longitudinal field studies of bartonella-infected rodent communities have generally failed to find correlations between flea abundance, flea infection rates, and rodent host infestation rates or infection prevalence (Telfer et al. Reference Telfer, Begon, Bennett, Bown, Burthe, Lambin, Telford and Birtles2007a; Morway et al. Reference Morway, Kosoy, Eisen, Montenieri, Sheff, Reynolds and Powers2008). Incongruences among these parameters may be explained by our study results which demonstrate that rodent hosts can have an extremely variable response in time to onset of bacteraemia. Differences in exposure dose and individual variation in susceptibility to infection in rodent populations may obscure our ability to find correlations among these parameters.
Bartonella bacteria were not detected in the urine of infected mice during this study although mice were bacteraemic at the time of sampling (weeks 3–6). Since urines were only collected for 4 weeks it is possible that additional sampling might have revealed the presence of bacteria in the urine later in the study. In addition, urines were frozen before testing, and this may have negatively impacted our ability to detect viable bacteria in the samples. However, in a previously published study urine collected from B. henselae-infected, bacteraemic cats was evaluated for the presence of viable bacteria or DNA without success (Kordick and Breitschwerdt, Reference Kordick and Breitschwerdt1997). This suggests that urinary excretion of bartonella bacteria is not common, if it occurs at all. However, because the relationship between specific bartonellae and their natural hosts is still not well understood, the possibility remains that this transmission mechanism could function in some natural reservoir populations.
The 2 B. tribocorum strains evaluated in this study share 100% nucleotide sequence identity for a portion of their citrate synthase gene (gltA), yet they exhibited different infection phenotypes in the laboratory mouse. This may be due to a number of factors. The most likely explanation for this phenomenon is naturally occurring variability in this bacterial strain population. It is possible that the differential infection phenotype exists in nature and functions to reduce competition for hosts between these phenotypic variants, but such an interaction would depend on a lack of cross-immunity between the strains.
Interestingly, strain MA appeared as rough colonies throughout the study when cultured from bacteraemic mice, whereas cultured ME produced consistently smooth colonies with some rough colonies present at times. This phenomenon, termed phase variation, has been described from rodent bartonella isolates previously (Kosoy et al. Reference Kosoy, Regnery, Tzianabos, Marston, Jones, Green, Maupin, Olson and Childs1997). In addition, rough/smooth colony phenotypes have been investigated for B. henselae and the rough colony morphology was shown to be associated with the expression of pili (Batterman et al. Reference Batterman, Peek, Loutit, Falkow and Tompkins1995). A rough colony producing B. henselae strain was 100 times more invasive in vitro than a smooth colony producing B. henselae strain (Batterman et al. Reference Batterman, Peek, Loutit, Falkow and Tompkins1995). Increased expression of pili by strain MA relative to strain ME could also explain differences in their in vivo infectivity for the mice.
In summary, we report on a mouse model for persistent bacteraemia using bartonella strains isolated from wild-caught Mus species. This system may be used to evaluate the transmission dynamics of bartonella bacteria among hosts or to investigate the molecular basis for host specificity demonstrated by some bartonella strains. Susceptibility of mice to infection at low bacterial doses, coupled with high bacteraemia levels make this system a superior candidate for vector transmission studies. Finally, the dynamics of bacterial co-infection and host competition can be explored using both MA and ME in the mouse model, potentially providing unique insights into bartonella bacterial ecology.