Introduction
Transgenic crops expressing toxins from Bacillus thuringiensis Berliner (Bt) have been used widely since 1996 to control key insect pests (Shelton et al., Reference Shelton, Zhao and Roush2002; James, Reference James2009). However, concern has been expressed that extensive and prolonged exposure to Bt toxins may select for resistance in target pest populations reducing the long-term utility of the technology. Understanding how resistance evolves is critical to developing effective resistance management programs that are necessary to sustain the technology (Gould, Reference Gould1988, Reference Gould1994, Reference Gould1998; Van Rie, Reference Van Rie1991; Roush, Reference Roush1994; United States Environmental Protection Agency, 2001; Tabashnik et al., Reference Tabashnik, Carrière, Dennehy, Morin, Sisterson, Roush, Shelton and Zhao2003).
The fall armyworm, Spodoptera frugiperda (J. E. Smith), is endemic to the Western Hemisphere and distributed from North America to Argentina (Sparks, Reference Sparks1979). It is an important pest of maize and cotton throughout the neotropics and a late season pest throughout the US (Wiseman & Davis, Reference Wiseman and Davis1979; Buntin, Reference Buntin1986; Mitchell et al., Reference Mitchell, McNeil, Westbrook, Silvain, Lalanne-Cassou, Chalfant, Pair, Waddill, Sotomayor-Rios and Proshold1991). S. frugiperda does not diapause and is vulnerable to freezing temperatures. Seasonal migrations to temperate regions of North America occur from overwintering populations in southern Florida, southern Texas and Mexico (Sparks, Reference Sparks1979; Buntin, Reference Buntin1986; Mitchell et al., Reference Mitchell, McNeil, Westbrook, Silvain, Lalanne-Cassou, Chalfant, Pair, Waddill, Sotomayor-Rios and Proshold1991). Population genetics studies suggest limited genetic exchange between Florida and Texas fall armyworm populations (Nagoshi et al., Reference Nagoshi, Meagher and Jenkins2010, Reference Nagoshi, Meagher and Hay-Roe2012). Texas populations migrate northward into Kansas, Nebraska, Iowa, Minnesota, Illinois and eastward to Pennsylvania. In contrast, migration from Florida appears to be limited to the southern Atlantic coastal states and is restricted to regions east of the Appalachian Mountain range. Overlap between Florida and Texas populations appears to occur in limited areas north and south of the primary elevations of the Appalachians (Nagoshi et al., Reference Nagoshi, Meagher and Hay-Roe2012). S. frugiperda exhibits two strains (maize and rice) based on host-plant preference. The maize strain feeds primarily on maize but also on cotton and sorghum. The rice strain feeds predominantly on rice, bermudagrass and johnsongrass (Pashley, Reference Pashley1986; Nagoshi & Meagher, Reference Nagoshi and Meagher2004). The strains are indistinguishable morphologically in larvae and adults (Pashley, Reference Pashley1988) but differ in their genetic constitution based on a number of different molecular markers (Pashley, Reference Pashley1986; Levy et al., Reference Levy, García-Maruniak and Maruniak2002; Nagoshi et al., Reference Nagoshi, Silvie, Meagher, López and Machado2007a), and in their physiology (Pashley, Reference Pashley1988; Quisenberry & Whitford, Reference Quisenberry and Whitford1988; Whitford et al., Reference Whitford, Quisenberry, Riley and Lee1988).
One of the more recent developments for managing fall armyworm populations has been the use of Bt transgenic maize, Zea mays L., expressing the Cry1F toxin (Siebert et al., Reference Siebert, Babock, Nolting, Santos, Adamczyk, Neese, King, Jenkins, McCarty, Lorenz, Fromme and Lassiter2008a, Reference Siebert, Tindall, Leonard, Van Duyn and Babcockb) that provides better control than hybrids producing Cry1Ab (Buntin et al., Reference Buntin, Lee, Wilson and McPherson2000; Stewart et al., Reference Stewart, Adamczyk, Knighten and Davis2001; Waquil et al., Reference Waquil, Ferreira Villela and Foster2002; Buntin, Reference Buntin2008; Hardke et al., Reference Hardke, Leonard, Huang and Jackson2011). Maize hybrids that express the Cry1F insecticidal protein from B. thuringiensis var. aizawai were developed by Dow AgroSciences (Indianapolis, IN) and Dupont Pioneer (Johnston, IA). These hybrids have been commercially available since 2003 and marketed as Herculex® I Insect Protection (transformation event TC1507). This product has demonstrated satisfactory control of S. frugiperda and other important lepidopteran pests (Siebert et al., Reference Siebert, Babock, Nolting, Santos, Adamczyk, Neese, King, Jenkins, McCarty, Lorenz, Fromme and Lassiter2008a, Reference Siebert, Tindall, Leonard, Van Duyn and Babcockb). Although TC1507 maize hybrids were commercialized in 2003, in Puerto Rico Cry1F expressing maize was first grown in 1998 for hybrid development and parental seed production as well as efficacy trials (Buntin, Reference Buntin2008). Unexpected damage to Cry1F maize hybrids was reported in 2006 in Puerto Rico and high levels of Cry1F resistance in fall armyworm was subsequently documented (Matten et al., Reference Matten, Head, Quemada, Romeis, Shelton and Kennedy2008; Tabashnik et al., Reference Tabashnik, Van Rensburg and Carrière2009; Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010).
Cry1F resistance evolution among S. frugiperda populations from Puerto Rico represents one of the few instances of documented field-evolved resistance to transgenic Bt crops. Other species with reported field-developed resistance include Busseola fusca (Fuller) resistant to Cry1Ab maize in South Africa (Van Rensburg, Reference Van Rensburg2007), Helicoverpa zea (Boddie) resistant to Cry1Ac and Cry2Ab cotton in southeastern US (Tabashnik et al., Reference Tabashnik, Van Rensburg and Carrière2009; Huang et al., Reference Huang, Andow and Buschman2011), Pectinophora gossypiella (Saunders) resistant to Cry1Ac cotton in India (Dhurua & Gujar, Reference Dhurua and Gujar2011) and Diabrotica virgifera vigifera LeConte resistant to Cry3Bb1 maize in Iowa, US (Gassmann et al., Reference Gassmann, Petzold-Maxwell, Keweshan and Dunbar2011). Field-evolved resistance of S. frugiperda to Cry1F maize occurred after only four years of commercialization, making this the fastest documented case of field-evolved resistance to a Bt crop and the first case of resistance leading to withdrawal of a Bt crop from the marketplace (Tabashnik et al., Reference Tabashnik, Van Rensburg and Carrière2009).
Storer et al. (Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010) confirmed that field-control failures of TC1507 maize in Puerto Rico were associated with a high-level of resistance. The highest Cry1F concentration tested against the resistant population (10,000 ng Cry1F per cm2) did not cause significant mortality, suggesting a resistance ratio in excess of 1000-fold. To evaluate inheritance, the F1 progeny from reciprocal crosses of the susceptible and resistant populations were bioassayed, and the dose–response statistics were compared. Mortality and growth inhibition data from the susceptible, resistant and F1 progeny were used to calculate dominance of resistance. The resistance to Cry1F was shown to be autosomal and highly recessive. Sensitivity of the resistant and susceptible colonies to Cry1Ab and Cry1Ac was also evaluated, and there was no indication of strong cross-resistance to these toxins (Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010, Reference Storer, Kubiszak, King, Thompson and Santos2012).
Multiple factors are thought to have contributed to the evolution of resistance to Cry1F in S. frugiperda in Puerto Rico (Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010, Reference Storer, Kubiszak, King, Thompson and Santos2012). Puerto Rico is an isolated island ecosystem that is subdivided by mountainous terrain. This terrain may restrict migration and dispersal and enable intense selection within local populations. In addition, the tropical environment of Puerto Rico allows year round cultivation of maize with multiple generations exposed to selection pressure in a calendar year. The long history of using formulated Bt insecticides for managing S. frugiperda in vegetables and seed maize, along with the use of other Bt maize events that produce Cry1Ab may have also contributed selection. The affected lines were silage hybrids, not adapted to tropical conditions and lacked native resistance traits (Storer et al., Reference Storer, Kubiszak, King, Thompson and Santos2012). Finally, although fall armyworm is highly polyphagous with many crop and non-crop hosts in Puerto Rico, in 2006 severe drought conditions forced fall armyworm populations to become concentrated in irrigated crops, of which Cry1F maize was an important component. The selection pressure in 2006 was likely to have been the most intense seen to date (Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010, Reference Storer, Kubiszak, King, Thompson and Santos2012). After resistance was reported in 2006, Storer et al. (Reference Storer, Kubiszak, King, Thompson and Santos2012) continued monitoring populations in Puerto Rico and in southern areas of the mainland US. The majority of collections from Puerto Rico continued to show high levels of Cry1F resistance whereas populations collected from the southern US have remained susceptible to Cry1F and TC1507 maize.
Although resistance to Cry1F has previously been characterized in a population from Puerto Rico (Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010), certain aspects of the resistance have yet to be addressed. In the present study, inheritance patterns of Cry1F resistance (dominance and number of loci) and cross-resistance to Cry1Aa, Cry1Ab, Cry1Ac, Cry1Ba, Cry2Aa and Vip3Aa were determined. A complete characterization of the inheritance of resistance allowed performing F1 screenings to detect the frequency of resistant alleles in field populations outside of Puerto Rico (Florida and Texas). The results of this research have direct implications for S. frugiperda resistance management for Cry1F maize.
Materials and methods
Insect strains and rearing
The Cry1F-selected strain was generated by Dupont Pioneer (Johnston, Iowa) and originated from several hundred field collected fall armyworm egg masses from Puerto Rico maizefields during October 2008 and January 2009. Egg masses were brought into the laboratory in Johnston, Iowa, where 826 neonates were selected by exposing them to TC1507 leaf discs. Only larvae that survived a 4-day exposure (785 larvae) were maintained and used to establish the Cry1F-selected strain. The susceptible strain was purchased from BioServ (Frenchtown, New Jersey), and has been in continuous culture since November 1997 with regular screenings to monitor for any changes in insecticide susceptibility. The BioServ strain, Cry1F-selected strain and field-collected larvae from Puerto Rico were identified as maize-strain. Strain identification was performed using a PCR amplification of a region of the mitochondrial COI gene with posterior digestion with EcoRV as described by Nagoshi et al. (Reference Nagoshi, Silvie and Meagher2007b).
Both strains were maintained using rearing techniques adapted from Perkins (Reference Perkins1979) with at least 200 adults randomly mating at each generation. Adults were placed in 31×23 cm wired hermit crab cages (Florida Marine Research, Sarasota, Florida) with diet placed in a cotton pad inside of the bottom of a 100×15 mm Petri dish (Fisherbrand, Waltham, Massachusetts) and replenished daily. Adult diet consisted of stale beer, ascorbic acid, propionic acid and aureomycin (Perkins, Reference Perkins1979). Adults were allowed to mate and lay eggs on wax paper. Eggs were harvested daily and placed in 100×15 mm Petri dishes with moistened filter paper until hatching. Larvae were reared on multispecies lepidopteran diet (BioServ, Frenchtown, New Jersey). Neonates were placed on shredded diet and allowed to grow until third instar. Approximately 300 third-instar larvae were individually transferred to 1 oz. translucent polystyrene soufflé portion cups (Solo Cup Company, Lake Forest, Illinois) with 4.5 ml of diet to minimize cannibalism. Pupation and adult emergence occurred within the cups. Emergent adults were transferred daily to mating cages.
Bt toxins
The Cry1F used in diet bioassays was expressed in BtG8 cells grown in CYS2 media with tetracycline and grown for six days at 30 °C. Cells were harvested by centrifugation and the pellets were washed five times with 0.5 M NaCl and twice with water. Washed pellets were stored at −20 °C. Pellets were lysed with 50 mM sodium carbonate pH 10, 10 mM DTT overnight at 4 °C. The lysate was concentrated with Millipore (Billerica, Massachusetts) concentrator devices (100,000 molecular weight cut off, MWCO) to ∼12.5 mg ml−1 and dialyzed against 50 mM Na carbonate/Na bicarbonate pH 10 using 25 K MWCO dialysis tubing. Aliquots of 5 mg and 20 mg were made, which were flash frozen in LN2, and then lyophilized.
Cry toxins used for cross-resistance experiments were prepared from fermentation of recombinant Escherichia coli (Migula) strains transformed to express Cry1Aa (ECE52), Cry1Ab (ECE53), Cry1Ac (ECE54), Cry1Ba (ECE128) and Cry2Aa (ECE126). The strains were obtained from the Bacillus Genetic Stock Center of The Ohio State University (Columbus, Ohio). Recombinant E. coli cultures were grown at 37 °C for 48 h in Luria-Bertani media. Protoxins were obtained from E. coli fermentation products following the method described by Lee et al. (Reference Lee, Milne and Dean1992). Toxin preparations were quantified by densitometric quantification (Crespo et al., Reference Crespo, Spencer, Nekl and Siegfried2008) of the 60–65 kDa peptides after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and compared to a standard curve for bovine serum albumin (BSA). Endotoxins were stored at −80 °C (Tan et al., Reference Tan, Cayabyab, Alcantara, Ibrahim, Huang, Blankenship and Siegfried2011).
Vip3A used was from a single source of lyophilized Vip3Aa19 protein supplied by Syngenta Biotechnology (Research Triangle Park, North Carolina). Vip3A protein was produced through an E. coli expression system and the protein was purified by anion exchange chromatography prior to lyophilization. Purity was assessed/quantified by Sypro Orange-stained SDS–PAGE. Lyophilized protein was kept frozen until use.
Bioassays
Bioassays were performed based on the methods described by Marçon et al. (1999) in 128-well bioassay trays (CD International, Pitman, New Jersey). One ml of European corn borer wheat germ-based diet (Lewis & Lynch, Reference Lewis and Lynch1969) was dispensed into each well and allowed to solidify. Seven concentrations of the toxin were used for LC50 determinations. Dilutions were made in 0.1% Triton-X 100 non-ionic detergent to obtain uniform spreading on the diet surface. Each well was surface treated by applying 30 μl of the appropriate concentration. The negative control consisted of wells treated with 30 μl of 0.1% Triton-X 100.
The treated wells were allowed to air dry, and one randomly selected S. frugiperda neonate (<24 h after hatching) was transferred using a fine paint brush into each well. Wells were covered with vented lids (BioServ, Frenchtown, New Jersey), and trays were held in an incubator at 27 °C, 24-h scotophase, and 80% RH. Mortality and combined larval weights were recorded seven days after infestation. Larvae that had not grown beyond first instar and weighed ≤0.1 mg were considered dead. Thus, mortality in this study includes both severe growth inhibition and death. Control mortality averaged 6% across treatments, and any replicates that exceeded 20% were not included. In each experiment, bioassays were replicated four times for each strain or cross, with 16 larvae per concentration (total of at least 64 larvae per concentration per cross).
Statistical analysis
Concentration–mortality data were analyzed by probit analysis (Finney, Reference Finney1971) using POLO-PC (LeOra Software, 1987). LC50 and LC99 were calculated, together with their 95% confidence intervals (CI), slopes and standard errors. A likelihood ratio test was conducted to test that the LC50s were equal. Larval weights were transformed to percent growth inhibition relative to the controls and these data were analyzed by non-linear regression using PROC NLIN (SAS Institute, 2011). Inverse regression was used to estimate GIC50 (effective concentration at which 50% growth inhibition level is attained), their 95% CI, slopes and standard errors. The diagnostic concentration was determined based on the upper 95% confidence limit (CL) of the LC99 of the susceptible strain, then confirmed in separate bioassays with the susceptible and resistant strains (Marçon et al., Reference Marçon, Siegfried, Spencer and Hutchison2000).
Sensitivity ratios were calculated using concentration–response statistics based on either mortality or growth inhibition. These values were calculated as the LC50 or GIC50 of the resistant strain divided by the LC50 or GIC50 of the susceptible strain (Robertson et al., Reference Robertson, Preisler, Ng, Hickle and Gelernter1995, Reference Robertson, Russel, Preisler and Savin2007). When mortality was not generated and growth inhibition was not higher than 50% at the highest concentration used, the highest concentration was utilized to calculate the sensitivity ratio. Sensitivity ratios were regarded as equal if the 95% CI of the estimate of these values did not overlap.
Inheritance experiments
To evaluate sex linkage and dominance of resistance, F1 progeny from reciprocal crosses between resistant and susceptible strains (susceptible ♀×resistant ♂ and susceptible ♂×resistant ♀) were bioassayed as previously described and mortality curves were produced. Sex linkage was determined using hypothesis tests to compare the slopes and intercepts of probit regressions derived from reciprocal crosses and parental strains. We tested the null hypothesis that the lines are neither parallel nor equal using POLO-PC (LeOra Software, 1987; Robertson et al., Reference Robertson, Russel, Preisler and Savin2007). Dominance of resistance was calculated using the method of effective dominance at a fixed concentration:
where X SS, XRS and X RR are the quantitative values calculated for a trait X for susceptible homozygotes, heterozygotes and resistant homozygotes, respectively. Values of D X range from zero or completely recessive resistance, to one representing completely dominant resistance. When D X is 0.5, resistance is referred as codominant or additive (Bourguet et al., Reference Bourguet, Genissel and Raymond2000). The traits used in the calculation of dominance were mortality (D ML) and growth inhibition (D GIL). D ML and D GIL were calculated at 7200 ng cm−2 which was the rate used in the calculations because it was the highest concentration tested. Mortality and growth inhibition were 100% at 7200 ng cm−2 in the susceptible strain, but no measurable effect was generated on the resistant strain. Estimates of D LC (based on LC50) could not reliably be assessed because mortality did not occur at the highest concentration in the resistant population and LC50 values could not be calculated (Bourguet et al., Reference Bourguet, Genissel and Raymond2000; Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010).
To estimate the number of loci that confer resistance to Cry1F, F1 progeny from reciprocal crosses were backcrossed to the resistant strain. The power of indirect tests for mode of inheritance is higher when the backcross progeny originate from crosses between the F1 progeny and the parental strain most dissimilar in susceptibility (Roush & Daly, Reference Roush, Daly, Roush and Tabashnik1990; Tabashnik, Reference Tabashnik1991). The monogenic inheritance model was tested using a χ2 test (Georghiou, Reference Georghiou1969; Preisler et al., Reference Preisler, Hoy and Robertson1990; Tabashnik, Reference Tabashnik1991, Tabashnik et al., Reference Tabashnik, Schwartz, Finson and Johnson1992). If resistance is monogenic, a backcross will produce progeny that are 50% RS and 50% RR. To test this hypothesis, the expected mortality in the backcross progeny at toxin concentration x was calculated using the formula
where W RS and W RR are the mortalities of the presumed RS (F1) and RR (parental line) genotypes at concentration x, respectively interpolated from probit regression. A χ2 goodness of fit test was conducted to determine differences between the observed and expected mortality of the backcross response and expected response at each concentration (Tabashnik, Reference Tabashnik1991; Tabashnik et al., Reference Tabashnik, Schwartz, Finson and Johnson1992).
Cross-resistance
To determine if Cry1F resistance in S. frugiperda caused changes in susceptibility to other Bt toxins, the susceptible and resistant strains were bioassayed against Cry1Aa, Cry1Ab, Cry1Ac, Cry1Ba, Cry2Aa and Vip3Aa proteins. The same bioassay methodology described above was used for all bioassays. LC50s were calculated for Cry1Ab and Vip3Aa using POLO-PC (LeOra Software, 1987). GIC50s were estimated for Cry1Ab, Cry1Ac and Vip3Aa with PROC NLIN (SAS Institute, 2011). Sensitivity ratios were generated for these toxins as previously explained.
Both strains showed limited response to Cry1Aa, Cry1Ba and Cry2Aa. Therefore, the highest achievable concentration was used for bioassays and individual larval weights were recorded (64 larvae per strain). Larval weights were transformed to percent growth inhibition relative to the control, and an analysis of variance was used to identify significant differences in inhibition between the susceptible and resistant strains using pairwise comparisons for each toxin using PROC GLIMMIX (SAS Institute version 9.2.2, 2011).
Frequency of resistant alleles
An F1 screen was used to identify the frequency of resistant alleles in field populations from areas where overwintering fall armyworm populations are known to occur (Texas and Florida). An F1 screen involves crossing field-collected individuals of unknown genotype with individuals from a resistant laboratory strain. The offspring was then bioassayed to allow discrimination among resistant homozygotes, susceptible homozygotes and heterozygotes (Gould et al., Reference Gould, Anderson, Jones, Summerford, Heckel, Lopez, Micinski, Leonard and Laster1997; Mahon et al., Reference Mahon, Downes, James and Parker2010).
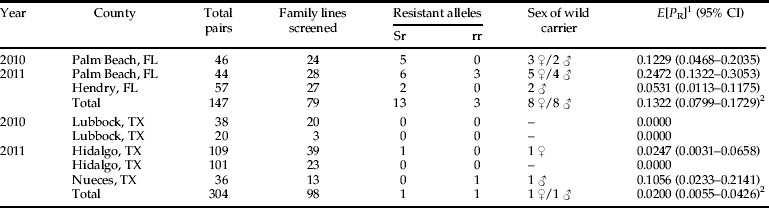
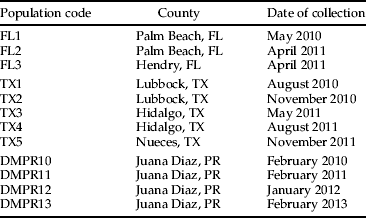
S. frugiperda populations were collected in Florida and Texas in 2010 and 2011 (table 1). Immature insects, eggs and larvae from field maize were reared on artificial diet and allowed to pupate as previously described. Pupae were sexed and individually paired with one or two individuals of the opposite sex from the resistant laboratory strain. Adults were placed in ‘honeymoon cages’ made of 27-gauge woven hardware cloth with a 33 mm diameter disposable plastic Petri dish (Sterilin, Newport, UK) used as bottom and top. The cages were 4.2 cm tall. Each cage had an opening on the top where a cotton ball saturated with adult diet was placed. To prevent diet dehydration, water was added every day. Wax paper was placed around the cage to provide an oviposition substrate. Eggs from each pair were collected daily and allowed to hatch. At least 48 neonates per pair were bioassayed with a Cry1F diagnostic concentration (200 ng cm−2) as previously described. Pairs were considered successful when they produced enough hatched neonates to be tested.
Table 1. Collections of S. frugiperda from Florida and Texas used for F1 analysis and from Puerto Rico to evaluate sensitivity to Cry1F.
The expected mortality at the diagnostic concentration is dependent on the genotype of the field-collected parent. If the field-collected parent is homozygous for susceptibility, the resulting progeny should all be heterozygotes resulting in 100% mortality at the diagnostic concentration. However, if the field collected parent carries one resistant allele a 1:1 ratio of heterozygotes to resistant homozygotes will result and approximately 50% mortality at the diagnostic bioassays would be expected. If the parent is homozygous for resistance, all progeny will be resistant and 100% survival at the diagnostic bioassay is expected (Gould et al., Reference Gould, Anderson, Jones, Summerford, Heckel, Lopez, Micinski, Leonard and Laster1997; Mahon et al., Reference Mahon, Downes, James and Parker2010). Larvae from the families that were identified from the F1 screen as being resistant were pooled, reared to adults and sib-mated. F2 offspring from these families were tested with the diagnostic concentration to confirm the presence of resistant alleles (Gould et al., Reference Gould, Anderson, Jones, Summerford, Heckel, Lopez, Micinski, Leonard and Laster1997).
Information from the F1 screenings was used to estimate resistance allele frequencies (E[P R]). For each collection the Bayesian methods described by Yue et al. (Reference Yue, Huang, Leonard, Moore, Parker, Andow, Cook, Emfinger and Lee2008) were used to estimate frequencies and 95% credible intervals for these estimates were obtained from equation (15) of Andow & Alstad (Reference Andow and Alstad1999). To calculate the probability of a false negative (P N0) in an F1 screen, equation (5) of Wenes et al. (Reference Wenes, Bourguet, Andow, Courtin, Carré, Lorme, Sanchez and Agustin2006) was used. Differences between Florida and Texas in total frequency of resistant alleles were analyzed using Fisher's exact test.
To test the prevalence of resistant alleles in Puerto Rico, field collected fall armyworm eggs were obtained from Puerto Rico in 2010, 2011, 2012 and 2013 (table 1). Eggs were allowed to hatch and neonates were used for bioassays with the diagnostic concentration (200 ng cm−2). Frequency of resistant alleles was calculated using the Hardy–Weinberg frequency of homozygotes (q 2=√q), assuming that the genotypes are in Hardy–Weinberg proportions (Falconer & Mackay, Reference Falconer and Mackay1996). Proportion of survival and frequency of resistant alleles between years was analyzed using a χ2 test for homogeneity.
Results
Resistance levels
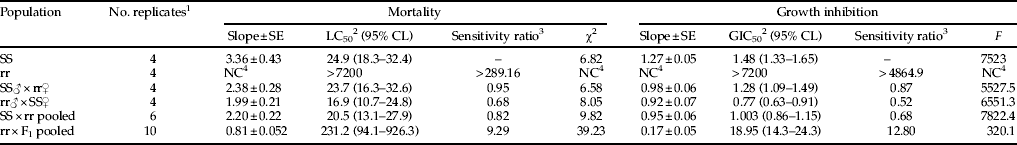
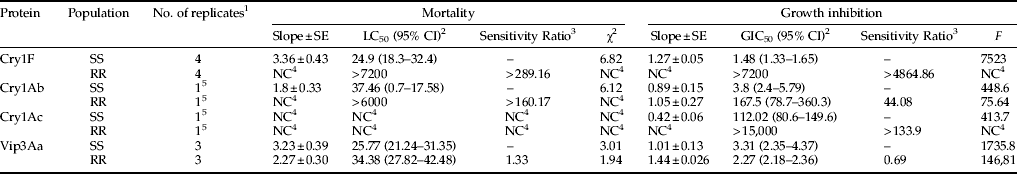
Bioassays revealed that the resistant laboratory strain displayed a high level of resistance to Cry1F. The LC50 for the susceptible strain was 24.86 ng cm−2 and for the resistant strain was greater than 7200 ng cm−2 (table 2), which was the highest concentration of Cry1F used in bioassays of this strain. This concentration was used to estimate the sensitivity ratio, indicating that the resistant population displays >289-fold resistance to purified Cry1F (table 2) and represents a conservative estimate of the sensitivity ratio. The diagnostic concentration was calculated to be 200 ng cm−2 based on the upper 95% CL of the LC99. The GIC50 for the susceptible strain was 1.48 ng cm−2 and for the resistant strain was again more than 7200 ng cm−2. The sensitivity ratio for growth inhibition was higher than 4865-fold (table 2).
Table 2. Concentration–response to Cry1F of resistant (rr), susceptible (SS), reciprocal crosses and backcrosses of S. frugiperda to Cry1F protein overlaid on artificial diet, as measured by both mortality (LC50) and growth inhibition (GIC50).

1 Each replicate consisted of 16 insects at each of the seven concentrations of Cry1F protein.
2 Nanograms of Cry1F per cm2 diet.
3 LC50 or GIC50 relative to the susceptible strain.
4 NC, not calculated because of insufficient dose response.
Inheritance of resistance
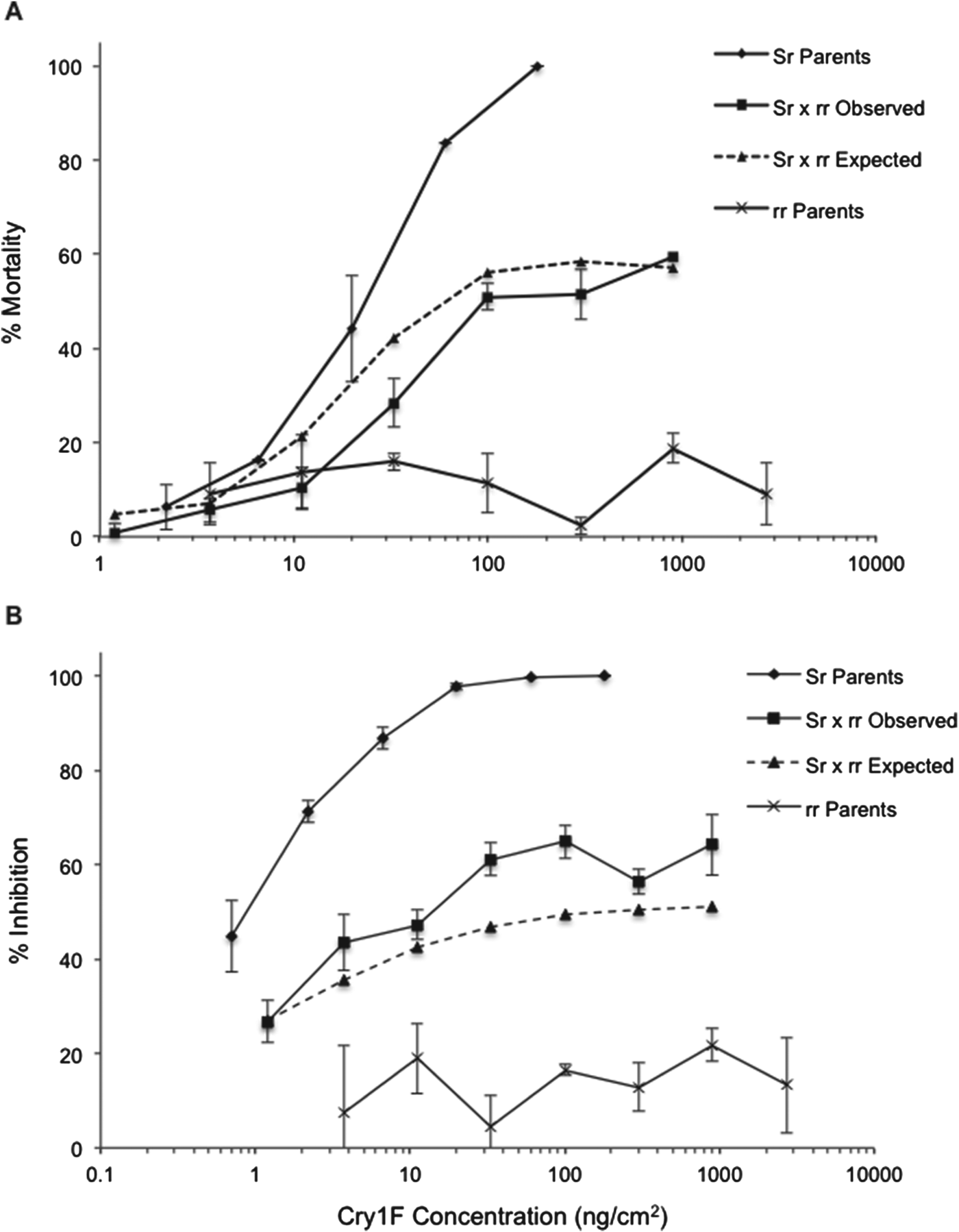
LC50s, GIC50s and sensitivity ratios of reciprocal crosses and backcrosses are presented in table 2. Analyses of mortality curves from the reciprocal crosses indicated that resistance to Cry1F in S. frugiperda is recessive and autosomal (fig. 1). The hypothesis of slope equality for mortality between the reciprocal crosses indicated that the slopes and intercepts are identical (χ2=5.33; df=2; P>0.05), confirming that the resistance is autosomal (Robertson et al., Reference Robertson, Russel, Preisler and Savin2007). Dominance was also examined by comparing the mortality response curves of the F1 generation with the most similar parental strain, in this case the susceptible strain. The test of equality showed no differences between the slopes and intercepts (χ2=9.02; df=4; P>0.05), indicating that resistance to Cry1F is recessive. The calculations of D ML and D GIL generated values of 0, confirming that resistance is recessive (table 3).

Fig. 1. Concentration–response curves of susceptible (SS), resistant (rr) and progeny of reciprocal crosses of S. frugiperda to Cry1F protein. Each point represents mortality (A) and growth inhibition (B) observed in four replications (see table 2) corrected for control mortality. Error bars represent standard errors of the mean mortality or inhibition at each concentration.
Table 3. Effective dominance estimates (D) for the Cry1F resistance trait in S. frugiperda from Puerto Rico compared with laboratory susceptible population. Mortality and growth inhibition measured at 7200 ng cm−2.

We also tested the monogenic versus polygenic inheritance model by backcrossing the F1 generation with the resistant strain (RS×RR) and comparing the progeny's response to the parent strains. The response curve of the backcross showed a plateau at 50% mortality (fig. 2), suggesting a 1:1 ratio of RS and RR genotypes. The direct test for monogenic inheritance showed no significant deviation between the observed and expected mortality at five of seven concentrations (table 4). At 11 and 33 ng cm−2, however, observed mortality was significantly lower than expected mortality (table 4), probably generated by genetic variance in the backcross progeny compared with the parental strains and the F1 (Tabashnik et al., Reference Tabashnik, Liu, Dennehy, Sims, Sisterson, Biggs and Carrière2002). As most of the concentrations tested were non-significant, resistance to Cry1F in S. frugiperda appears to be monogenic.

Fig. 2. Concentration–response curves of the backcross progeny compared with those of the F1 (rS) and the resistant parents (rr). Each point represents mortality (A) and growth inhibition (B) observed in four replications (see table 2) corrected for control mortality. Error bars represent the standard errors of the mean mortality or inhibition at each concentration.
Table 4. Direct test for deviation between the observed and expected mortality for a monogenic model (df=1).
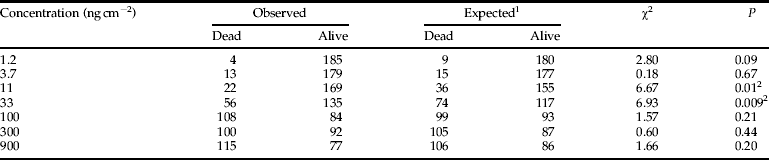
1 Expected % mortality at each concentration x, calculated as: Y x=0.5(% mortality of F1 at x+% mortality of R×S (pooled)).
2 Probability values indicating significant differences between the observed and expected mortality (P<0.05).
Cross-resistance
Cry1Ab and Cry1Ac were the only Cry proteins that generated a measurable response that allowed comparisons of the susceptible and resistant strains. Cry1Ab was the only Cry toxin with sufficient mortality in the susceptible strain to calculate an LC50. The LC50 for the susceptible strain was 37.46 ng cm−2. In comparison, the resistant strain had no mortality at 6000 ng cm−2. The GIC50 were 3.8 and 167.5 ng cm−2 for the susceptible and resistant strain, respectively. Sensitivity ratios for Cry1Ab were >160.17 times for mortality and 44.08 for growth inhibition (table 5). The hypothesis of equality was rejected (χ2=74.11; df=2; P>0.05), whereas the hypothesis of parallelism was not (χ2=1.27; df=1; P>0.05), indicating that intercepts are different, but slopes are equal (Robertson et al., Reference Robertson, Russel, Preisler and Savin2007). For Cry1Ac, the susceptible strain did not exhibit significant mortality at any of the concentrations tested, although significant growth inhibition was observed. The GIC50 for the susceptible strain was 112.02 ng cm−2 and for the resistant strain >15,000 ng cm−2, the highest concentration tested. The calculated sensitivity ratio for growth inhibition in Cry1Ac was >133.9 (table 5). Equal slopes with different intercepts indicate that for Cry1Ab and Cry1Ac both strains (resistant and susceptible) have qualitatively identical, but quantitatively different mortality responses (Robertson et al., Reference Robertson, Russel, Preisler and Savin2007). Results from Cry1Ab and Cry1Ac bioassays suggest a low level of cross-resistance relative to Cry1F in the resistant S. frugiperda from Puerto Rico.
Table 5. Comparative susceptibility of S. frugiperda susceptible and resistant strains to Cry1F, Cry1Ab, Cry1Ac and Vip3Aa.

1 Each replicate consisted of 16 insects at each of the seven concentrations of protein.
2 Nanograms of protein per cm2 diet.
3 LC50 or GIC50 relative to the susceptible strain.
4 NC, not calculated because of insufficient dose response.
5 Data collected from individual weights of larvae.
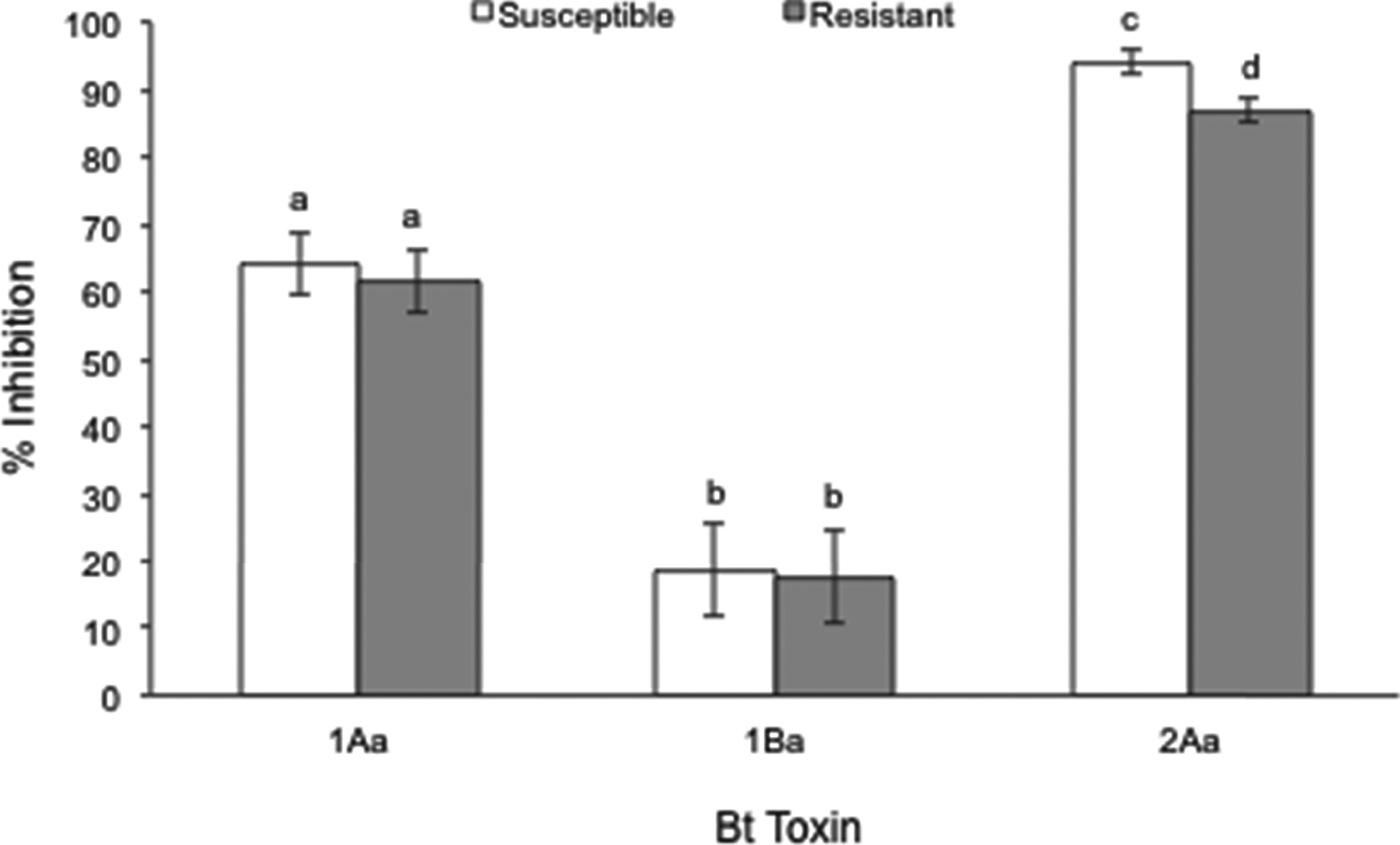
Responses of the susceptible and resistant strains to Cry1Aa, Cry1Ba and Cry2Aa were tested but neither strain showed significant mortality or growth inhibition at the highest achievable concentration (fig. 3). The Cry1F resistant strains exhibited higher growth inhibition when exposed to Cry2Aa (t=−4.10; df=1; P<0.0001). These results suggest that there is no cross-resistance with Cry1Aa and Cry1Ba. Significant differences in the response of the susceptible and the resistant colony to Cry2Aa could be indicating a slight level of negative cross-resistance between Cry2Aa and Cry1F. However, this difference might also be explained by natural variation among populations (fig. 3).

Fig. 3. Percentage of inhibition produced by the highest concentration of Cry1Aa (15,000 ng cm−2), Cry1Ba (12,000 ng cm−2) and Cry2Aa (5000 ng cm−2) in the susceptible and resistant strains. Error bars represent the standard errors of the mean inhibition. Bars with the same letter are statistically similar (t test, P>0.05).
Vip3Aa bioassays resulted in a similar response from both strains. The susceptible strain exhibited an LC50 of 25.77 ng cm−2 and the resistant strain of 34.38 ng cm−2. GIC50s were 3.31 ng cm−2 for the susceptible strain and 2.27 ng cm−2 for the resistant strain (table 5). The hypothesis of equality for mortality between the susceptible and the resistant strain indicated that the slopes and intercepts are identical (χ2=5.5; df=2; P>0.05), suggesting that there is no cross-resistance between Vip3Aa and Cry1F.
Frequency of resistant alleles
F1 results to detect frequency of Cry1F resistant alleles in populations of S. frugiperda from Florida and Texas are presented in table 6. Resistant alleles were more frequent for Florida than for Texas in both years. Five heterozygotes were found in Palm Beach County, Florida in 2010 representing an estimated resistant allele frequency of 0.1229. In 2011 six heterozygotes and three homozygote resistant individuals were identified in Palm Beach County, Florida resulting in an estimated allele frequency of 0.2472. Two heterozygotes were found in Hendry County, Florida with a subsequent estimated allele frequency of 0.0531. Although these locations are only 70 miles apart, differences in the frequency of resistant alleles were found.
Table 6. Frequency of Cry1F resistant alleles in S. frugiperda populations from Florida and Texas in 2010 and 2011.

1 Resistant allele frequency.
2 Resistant allele frequency E[P R] in Florida is significantly different from Texas (Fisher's exact test, P<0.0001).
No resistant alleles were found in Texas in 2010. In 2011 one homozygote resistant was found in Hidalgo County, resulting in an estimated allele frequency of 0.0247. One heterozygote was found in Nueces County in 2011 resulting in an estimated allele frequency of 0.1056. There were no differences in the sex of the wild carrier of the resistant allele for both Florida and Texas populations (table 6). Florida's total frequency of resistant alleles for 2010 and 2011 was 0.1322, while for Texas was 0.02, Fisher's exact test indicated significant differences between states (P<0.0001) (table 6). When control mortality is 10% and the total number of F1 larvae entering the screen is 30, the probability of finding a false negative for a line (P No) was 1.6×10−8 suggesting a very high detection probability (Wenes et al., Reference Wenes, Bourguet, Andow, Courtin, Carré, Lorme, Sanchez and Agustin2006).
Bioassays performed on insects from Puerto Rico maize in 2010, 2011, 2012 and 2013 indicated that proportion of survival and frequency of resistant alleles varied between years (χ2=44.92; P<0.0001). Regardless of fluctuations between years, high levels of resistant alleles remained constant for four years (table 7).
Table 7. S. frugiperda populations from Juana Diaz, Puerto Rico tested for sensitivity to Cry1F protein on artificial diet tested in 2010, 2011, 2012 and 2013.
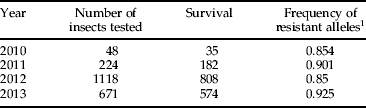
1 Frequency of resistant alleles was calculated using Hardy–Weinberg frequency of homozygotes (q 2=√q).
Proportion of survival and frequency of r alleles did not vary significantly between years (χ2 test for homogeneity=44.92, P<0.0001).
Discussion
The present study confirms the results reported by Storer et al. (Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010) in which S. frugiperda populations from Puerto Rico were highly resistant to Cry1F compared to a laboratory-susceptible population. Initial genetic characterization of resistance indicated that resistance was autosomal with no maternal effects, and highly recessive (Storer et al., Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010). Our results also indicate that resistance is autosomal and highly recessive based on both growth inhibition and mortality response curves in diet bioassays. In addition, bioassays of progeny resulting from crosses of the resistant parental strain to heterozygotes indicate that resistance to Cry1F in S. frugiperda is conferred by a single locus, which has not been previously reported.
Cross-resistance experiments suggest that there is a low level of cross-resistance to Cry1Ab and Cry1Ac, although the level of resistance is much lower than observed for Cry1F. These results are important to assist in identifying possible mechanisms of resistance and to guide decisions on which toxins are compatible for pyramided events. Storer et al. (Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010) reported similar results with Cry1Ac, but lower levels of cross-resistance with Cry1Ab. The Cry1F resistance ratios based on mortality and growth inhibition found here differed from Storer et al. (Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010), although similar trends were observed. Discrepancies in levels of cross resistance between the two studies might be the result of differences in the methodology, origin of the Cry proteins and/or the populations tested. Populations used in this study were collected at different times and locations compared to those used by Storer et al. (Reference Storer, Babcock, Schlenz, Meade, Thompson, Bing and Huckaba2010), and it is known that S. frugiperda response to Cry1A proteins is variable across geographies (Monnerat et al., Reference Monnerat, Martins, Queiroz, Ordúz, Jaramillo, Benintende, Cozzi, Real, Martinez-Ramirez, Rausell, Cerón, Ibarra, del Rincón-Castro, Espinoza, Meza-Basso, Cabrera, Sánchez, Soberón and Bravo2006). Cross-resistance among Cry1F, Cry1Ac and Cry1Ab suggests that altered midgut receptors could be responsible for resistance to Cry1F in S. frugiperda. Receptor binding studies with S. frugiperda and other Lepidoptera suggest Cry1A proteins share a common binding site with Cry1F (Luo et al., Reference Luo, Banks and Adang1999; Ferré & Van Rie, Reference Ferré and Van Rie2002; Hernández-Martínez et al., Reference Hernández-Martínez, Ferré and Escriche2009).
Results of bioassays with Cry1Aa, Cry1Ba and Cry2Aa indicate that fall armyworm is generally insensitive to these proteins, although some growth inhibition was observed at high doses. Cry1Aa and Cry1Ba showed no significant differences between the resistant and susceptible strains, indicating that there is no cross-resistance between these toxins and Cry1F. Although susceptibility to Cry2Aa was significantly higher for the resistant strain, the difference was slight and the suggestion of negative cross-resistance is uncertain and may reflect natural variation in susceptibility between populations. Finally, Vip3Aa bioassays suggest that there is no cross-resistance between Cry1F and Vip3Aa. This result supports the binding experiments that suggest a lack of competitive binding between Cry1F and Vip3A (Sena et al., Reference Sena, Hernandez-Rodriguez and Ferré2009). These results suggest the high potential of Vip3Aa to control Cry1F-resistant S. frugiperda and for the two toxins to be deployed as pyramided toxins.
The nature of Cry1F resistance inheritance (i.e., autosomal, recessive and conferred by a single locus) provides an efficient tool to detect resistance alleles among field populations using an F1 screening approach. Results of these tests suggest that the Cry1F resistance allele detected in both Florida and Texas is the same as that observed in Puerto Rico. Based on these results, the frequency of resistant alleles in Florida can be as high as 13%, but localized differences may exist. The frequency of resistance among Texas populations was much lower, but still detectable (0.02). These results are consistent with gene flow studies where genetic exchange between Puerto Rico and Florida has been identified based on mitochondrial haplotype ratios, while there is limited genetic exchange between Florida and Texas (Nagoshi et al., Reference Nagoshi, Meagher and Jenkins2010, Reference Nagoshi, Meagher and Hay-Roe2012). Migration of resistant individuals from Puerto Rico to Florida might be playing an important role in the higher frequency of resistant alleles in southern Florida, but local selection may also be affecting frequency estimates.
Prior selection pressures from Bt foliar sprays, and/or local selection with Cry1F expressing maize may also be affecting the frequency of resistant alleles. Studies with Spodoptera exigua (Hübner) have shown that selection with a moderately effective Bt protein, such as Cry1Ab, can lead to decrease sensitivity to other more effective proteins such as Cry1F (Hernández-Martínez et al., Reference Hernández-Martínez, Ferré and Escriche2009). Because Cry1Ab maize is grown in Texas, Florida and Puerto Rico and given the evidence of the low cross-resistance of Cry1F with Cry1Ab, Cry1F resistance allele frequencies may be influenced by Cry1Ab exposure. Unfortunately, it is difficult to determine the amount of Cry1F expressing maize grown in southern Florida. Local differences in frequencies between counties in Florida may be a result of differences in selection pressures with some areas having a greater production of Cry1F expressing maize. Additional studies are necessary to ultimately define the factors influencing the differences in frequency of resistant alleles between Florida and Texas, and local differences that may exist in southern Florida.
Results from discriminating bioassays from insects collected from Juana Diaz, Puerto Rico during 2010, 2011, 2012 and 2013 are similar to those reported by Storer et al. (Reference Storer, Kubiszak, King, Thompson and Santos2012) who also tested a collection from that municipality. Neither growth inhibition nor mortality reached 90% at the highest Cry1F concentrations tested. Our results with the diagnostic bioassay indicated the frequency of Cry1F resistance remains high although a low frequency of susceptible alleles may exist. The frequency of resistant alleles reported in this study may not reflect other local populations from Puerto Rico, where collections from Santa Isabel and Lajas populations exhibited a complete lack of response to Cry1F in 2010 and 2011, indicating the absence of susceptible alleles (Storer et al., Reference Storer, Kubiszak, King, Thompson and Santos2012).
Cry1F resistance in S. frugiperda is similar to the Cry1F laboratory-selected O. nubilalis in that inheritance of Cry1F resistance is autosomal, recessive and conferred by a single locus (Pereira et al., Reference Pereira, Lang, Storer and Siegfried2007, Reference Pereira, Storer and Siegfried2008). However, cross-resistance results differ slightly in that O. nubilalis exhibited low levels of cross-resistance to Cry1Ac and lack of cross-resistance to Cry1Ab, while S. frugiperda exhibited cross-resistance to both Cry1Ab and Cry1Ac (Pereira et al., Reference Pereira, Lang, Storer and Siegfried2007, Reference Pereira, Storer and Siegfried2008). Similarly, it has been suggested that the frequency of Cry1F resistant alleles in midwestern US, O. nubilalis populations may be higher than anticipated, and may have already been present at relatively higher frequencies prior to introduction of Cry1F-expressing plants (Siegfried, personal communication). Higher frequencies of Cry1F resistant O. nubilalis and S. frugiperda may suggest that there is a low fitness cost associated with Cry1F resistance in the absence of selection. Pereira et al. (Reference Pereira, Storer and Siegfried2009) reported that Cry1F resistant O. nubilalis are not significantly different from susceptible larvae of similar genetic background based on a number of parameters associated with reproductive fitness. A similar pattern might be occurring in S. frugiperda based on the relatively higher frequencies observed in Florida and the stability of resistance in Puerto Rico over a period of four years after Cry1F expressing plants are no longer commercially available. It is important to investigate in detail the existence of fitness costs associated with resistance. Comparisons of fitness traits, such as developmental time, fecundity and longevity in the susceptible and resistant strains, as well as the F1 progeny, will provide valuable information for resistance management and mitigation (Siegfried et al., Reference Siegfried, Zoerb and Spencer2001; Pereira et al., Reference Pereira, Storer and Siegfried2009; Crespo et al., Reference Crespo, Spencer, Tan and Siegfried2010). Preliminary results indicate that resistance to Cry1F in S. frugiperda is not associated with fitness cost (Vélez et al., unpublished).
Our results indicate that Cry1F resistance alleles are present in S. frugiperda populations in Florida and Texas, and US mainland populations are at risk for evolving field resistance. However, because resistance is recessive, its evolution may be delayed by compliance with refuge recommendations (Gould, Reference Gould1998). The current refuge requirement for single trait hybrids is 50% and 20% for pyramided products in the southern US in counties where both maize and cotton are grown. The refuge strategy combined with effective pyramided crops with multiple modes of action against S. frugiperda could help delay the spread of Cry1F resistant alleles (Adamczyk & Mahaffey, Reference Adamczyk and Mahaffey2008; Storer et al., Reference Storer, Kubiszak, King, Thompson and Santos2012). To date, there have been no reports of reduced effectiveness of Cry1F-expressing maize against S. frugiperda in Florida or Texas (Tabashnik et al., Reference Tabashnik, Van Rensburg and Carrière2009; Hardke et al., Reference Hardke, Leonard, Huang and Jackson2011; Storer et al., Reference Storer, Kubiszak, King, Thompson and Santos2012). Nonetheless, implementation of monitoring programs together with the investigation of reports of unexpected damage to Cry1F-expressing maize should be a priority. If reduction of product efficacy is linked to changes in allele frequency, actions should be taken to limit survival of resistant insects and slow or prevent their spread (Siegfried et al., Reference Siegfried, Spencer, Crespo, Storer, Head, Owens and Guyer2007). The use of insecticides when populations are high could also help reduce the frequency of resistant alleles (Storer et al., Reference Storer, Kubiszak, King, Thompson and Santos2012).
In order to have a better understanding of the evolution of resistance in S. frugiperda in Puerto Rico it is important to continue studying other aspects of the biology of this insect that could be affected by the presence of resistant alleles (e.g. fitness, behavior and migration). Further studies will help us to understand how resistance evolved in Puerto Rico and to predict future problems with this insect. Understanding field resistance will assist the development of better risk assessments, improve predictions of resistance to Bt crops in other Lepidoptera and maximize the benefits of current and future generations of transgenic crops. Information derived from Cry1F resistant S. frugiperda from Puerto Rico can guide resistant management strategies for Latin America where this insect is an important pest of maize and cotton. Planned deployment of Bt crops in Latin America suggests the need for resistant management programs designed for tropical areas where crop production is year round and pest pressure is continuous.
Acknowledgments
The authors thank Dupont Pioneer for providing the resistant fall armyworm strain from Puerto Rico and the Cry1F toxin used for bioassays. Syngenta provided Vip3Aa toxin. Dr Pat Porter, Dr Noel Troxclair and Monti Vandiver from Texas AgriLife Research and Extension for Texas collections of fall armyworm in 2010 and Taurino Trujillo, Jorge Guzman and Dr Marlin Rice from Dupont Pioneer for assisting with fall armyworm collections in 2011.