Introduction
The species complex Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) has a global distribution, and its members are major pests of agricultural crops, causing damage directly by feeding and indirectly by transmitting many plant viruses (Jones, Reference Jones2003). Their propensity to develop insecticide resistance, in combination with the high genetic variability, makes their control problematic. Although morphologically indistinguishable, members of this species complex display a large number of biological, physiological and genetic variations, which had led to the characterization of >30 biotypes (references in Xu et al., Reference Xu, De Barro and Liu2010). Recently, it has been proposed, based on a global analysis of mitochondrial cytochrome oxidase I gene (mtCOI) as well as the results of crossing experiments, that B. tabaci is a complex of at least 28 cryptic species (Dinsdale et al., Reference Dinsdale, Cook, Riginos, Buckley and De Barro2010; De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011; Hu et al., Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011 and references therein). These papers point out the erroneous and misleading use of the term biotype. Following their considerations, we use hereafter ‘Bemisia tabaci complex’ to refer to the species complex. In addition, we refer to the species corresponding to the clusters which have been determined by the Bayesian analysis of the mtCOI haplotypes, using the cluster's names proposed by Dinsdale et al. (Reference Dinsdale, Cook, Riginos, Buckley and De Barro2010) and adopted by Hu et al. (Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011). For example, we refer to the Middle East–Asia Minor 1 cluster (known commonly as biotypes B and B2) by using MEAM1. However, we refer to the species of the Mediterranean cluster (known commonly as biotypes Q, J and L) and which includes sequences from Greek whiteflies using Bemisia tabaci given that the lectotype of B. tabaci is from Greece. Greece, both mainland and insular regions, is the geographic origin of the whitefly populations of the present study, including the area of Agrinio in west Greece from which Gennadius (Reference Gennadius1889) first described the species. Moreover and in order to establish a connection with previous works, we continue to use the associated ‘biotype’ designation (B, Q) when appropriate.
Bemisia tabaci complex is known to harbour seven different vertically-transmitted bacteria (Zchori-Fein & Brown, Reference Zchori-Fein and Brown2002). Besides Portiera aleyrodidarum, the obligatory primary endosymbiont of whiteflies, which has a long co-evolutionary history with all members of the Aleyrodinae (Thao & Baumann, Reference Thao and Baumann2004), six additional facultative secondary endosymbionts were detected (Zchori-Fein & Brown, Reference Zchori-Fein and Brown2002). The interaction of Fritschea bemisiae, so far only reported in the New World species of the B. tabaci complex (Thao et al., Reference Thao, Baumann, Hess, Falk, Ng, Gullan and Baumann2003), with its host is as yet uncharacterized. Hamiltonella defensa, which induces parasitoid resistance in the pea aphid (Oliver et al., Reference Oliver, Degnan, Burke and Moran2010) seems to play a major role in virus transmission of MEAM1 (Gottlieb et al., Reference Gottlieb, Zchori-Fein, Mozes-Daube, Kontsedalov, Skaljac, Brumin, Sobol, Czosnek, Vavre, Fleury and Ghanim2010). Four others (Wolbachia, Cardinium, Rickettsia, Arsenophonus) are known to manipulate host reproduction in many different insect species (Hunter et al., Reference Hunter, Perlman and Kelly2003; Duron et al., Reference Duron, Bouchon, Boutin, Bellamy, Zhou, Engelstädter and Hurst2008; Saridaki & Bourtzis, Reference Saridaki and Bourtzis2010), but their effects on populations of the B. tabaci complex have yet to be determined. Rickettsia seems to play a role in the response to parasitization of the MEAM1 by the wasp Eretmocerus mundus (Mahadav et al., Reference Mahadav, Gerling, Gottlieb, Czosnek and Ghanim2008). Moreover, it has been shown that this bacterium increased the survival and fecundity of MEAM1 populations in the southwestern United States, leading to its spread in host populations in this area (Himler et al., Reference Himler, Adachi-Hagimori, Bergen, Kozuch, Kelly, Tabashnik, Chiel, Duckworth, Dennehy, Zchori-Fein and Hunter2011). These different species of symbionts often co-infect individuals of B. tabaci complex, and their community composition is closely related to the host species (Chiel et al., Reference Chiel, Gottlieb, Zchori-Fein, Mozes-Daube, Katzir, Inbar and Ghanim2007; Gueguen et al., Reference Gueguen, Vavre, Gnankine, Peterschmitt, Charif, Chiel, Gottlieb, Ghanim, Zchori-Fein and Fleury2010; Skaljac et al., Reference Skaljac, Zanic, Goreta Ban, Kontsedalov and Ghanim2010) although differences occur even within host species, as is the case for MEAM1 and B. tabaci (Gueguen et al., Reference Gueguen, Vavre, Gnankine, Peterschmitt, Charif, Chiel, Gottlieb, Ghanim, Zchori-Fein and Fleury2010).
MEAM1 (B biotype) and Q B. tabaci, both considered native to the Mediterranean Basin area, are highly invasive and currently occur worldwide (McKenzie et al., Reference McKenzie, Hodges, Osborne, Byrne and Shatters2009, Hu et al., Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011). The examination of the mtDNA polymorphism of Greek B. tabaci, both from insular and mainland Greece, had shown a high homogeneity of the COI sequences (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007). In addition, phylogenetic and diagnostic analysis disclosed that the Q and, more specifically, the western Mediterranean Q1 is predominant in Greece (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007; Roditakis et al., Reference Roditakis, Grispou, Morou, Kristoffersen, Roditakis, Nauen, Vontas and Tsagkarakou2009). The eastern Mediterranean Q2 has not been found in Greece, whereas the B seems to be restricted to some areas (Papayannis et al., Reference Papayiannis, Brownm, Hadjistylli and Katis2008).
Studies on the population genetics, using microsatellites, showed that both Q and B populations of the Mediterranean Basin are highly differentiated (Simón et al., Reference Simón, Cenis and De La Rúa2007). Microsatellite markers have been used to study the genetic structure of B and Ms populations of the southwest part of the Indian Ocean (Delatte et al., Reference Delatte, David, Granier, Lett, Goldbach, Peterscmitt and Reynaud2006, Reference Delatte, Holota, Warren, Becker, Thierry and Reynaud2011) and the characterization of at least six Asia-Pacific B. tabaci genetic populations with little or no gene flow between them (De Barro, Reference De Barro2005).
An exploratory study using only six B. tabaci samples from the island of Crete, Greece, showed that differentiation might be significant even at a small geographic scale, and the Bayesian approach disclosed that the individuals clustered into at least two groups based on their genotypes (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007). However, only six sampling populations, all from Crete, were included in that study, and any attempt to explain the factors affecting the observed structure would have led to speculations. To explore this issue, in the present study, we examined the polymorphism of eight microsatellite loci in a large set of sampling populations. By using populations coming from insular and continental Greece and collected on different plant species grown in both greenhouse and open environments, we addressed the role of the geographic distance, type of the habitat and host plant in shaping the genetic structure of the Q B. tabaci. In addition, with focus on Wolbachia, we characterized the bacterial symbiotic community known to induce reproductive alterations of host species, and thus possibly affecting (or affected by) the genetic structure of B. tabaci.
Material and methods
Whitefly samples
Samples were taken in mainland Greece (ten samples) and on the islands of Crete (26 samples), Santorini (one sample) and Schoinoussa (one sample) between 2002 and 2007 (fig. 1). A detailed record for each population is given in table 1. Of the total of 38 samples, seven were collected on non-cultivated plants (mainly Ipomoea sp. and Solanum nigrum) and 31 were from cultivated vegetables (mainly melon, cucumber, eggplant, pepper) or non-food crops (cotton and tobacco). Twelve samples, all from Crete, were from greenhouse plants.

Fig. 1. Geographical sites of B. tabaci collections. A unique identification code was assigned to each collected population and is used for reference along with a three-letter designation: MGr for samples from mainland Greece, SCr for samples from south Crete, NCr for north Crete, ISc for the island of Schoinoussa and ISa for the island of Santorini.
Table 1. Collection information and genetic characteristics of the populations.

N, number of individuals genotyped; N A, mean number of alleles per population; H O, observed heterozygosity; H E, expected heterozygosity; F IS, inbreeding coefficient; G, samples collected in greenhouses; F, samples collected from field cultivations. Significant Hardy-Weinberg departures after Bonferroni's correction are given in bold: *P<0.05, **P<0.01, ***P<0.001.
A unique identification code was assigned to each collected population and is used for reference along with a three-letter designation: MGr for samples from mainland Greece, SCr for samples from south Crete, NCr for north Crete, ISc for the island of Schoinoussa and ISa for the island of Santorini. Most of the Cretan samples were collected in the south, from the three main greenhouse crop regions, Ierapetra, Arvi and Tympaki, where pest management relies on intensive spraying of neonicotinoids, organophosphates, carbamates and pyrethroids (Roditakis et al., Reference Roditakis, Grispou, Morou, Kristoffersen, Roditakis, Nauen, Vontas and Tsagkarakou2009). At each location, adult whiteflies were collected from several plants within the same field (open environments or greenhouses) and stored until use at either −80°C or in 70% ethanol.
Genomic DNA was extracted from individual females as described previously (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007) and used for polymerase chain reactions (PCR).
Microsatellite genotyping
B. tabaci is haplo-diploid: males hatch from unfertilized eggs and are haploid. All genotyping was done using adult, diploid females. Each insect was genotyped at eight variable microsatellite loci. Details on the microsatellite markers used are given in the table 2. Two multiplex PCRs were performed for each individual. The first for loci BT-4, BT-b159 and BT-d26 and the second for loci BT-b34, BT-b155, 11, 145 and 177 with annealing for 45 s at 63°C and 59°C, respectively. One or two μl of DNA extract were used in each 10 μl PCR reaction, containing 0.2 mM dNTPs, 1.6 mM MgCl2, 1× enzyme buffer (Minotech) and 0.5 units Taq polymerase (Minotech). The final primer concentrations and fluorescent labels are shown in table 2. The PCR product was diluted 1/8, one μl from the dilution was mixed with 11 μl of Hi-Di™ formamide and 0.2 μl Genescan Liz 400 size standard (Applied Biosystems), denatured for 5 min at 96°C, and electrophoresed with an ABI 3700 sequencer. Micro-Checker v. 2.2.3 (Oosterhout et al., Reference Oosterhout, Hutchinson, Wills and Shipley2004) was used to evaluate the genotyping results for null-alleles and scoring errors.
Table 2. Characteristics of the microsatellite loci used in the population genetic study of Bemisia tabaci.

Locus BT-d26 was removed from further studies due to evidence for the presence of null alleles, see results for details.
A, number of alleles; H O, observed and H E, expected heterozygosity per locus. Loci BT-4, BT-b159, BT-d26, BT-b34, BT-b155, are from Tsagkarakou & Roditakis (Reference Tsagkarakou and Roditakis2003), locus 11 from Delatte et al. (Reference Delatte, David, Granier, Lett, Goldbach, Peterscmitt and Reynaud2006) and loci 145 and 177 are from Dalmon et al. (Reference Dalmon, Halkett, Granier, Delatte and Peterschmitt2008). Fluorochroms used for PCR product detection are indicated as Fam, Ned and Vic.
From most sampling sites, 20 to 33 individuals were genotyped, resulting in about 1000 individuals for each locus (table 1).
Species identification
Species identity was examined first through the allele size of microsatellite locus BT159, which has been shown to be diagnostic for the B and Q populations and through a PCR-RFLP test to further discriminate between the Q1 (west Mediterranean) and the Q2 (east) populations (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007). The latter assay was performed for all the individuals of the 19 populations included in the endosymbiont study.
Microsatellite data analysis
Linkage disequilibrium, departure from Hardy-Weinberg Equilibrium (HWE), inbreeding coefficient (F IS), expected (H E) and observed (H O) heterozygosities, as well as allele frequencies and mean number of alleles per locus were tested/calculated using software GENEPOP, version 4 (Rousset, Reference Rousset2007). When multiple tests were conducted, sequential Bonferroni correction of the P-values was performed (Holm, Reference Holm1979). The distributions of two groups of H or F IS estimates were compared by performing a Mann-Whitney U-test.
F ST estimates were computed according to Weir & Cockerham (Reference Weir and Cockerham1984). Genotypic differentiation between and among populations were analyzed by computing an unbiased estimate of the P-value of a F ST based exact test, as implemented in GENEPOP, version 4. The overall significance of multiple tests for each locus was estimated using the Fisher combined probability test. In addition to F-statistics which rely on a predefined population organisation, a model-based method (Pritchard et al., Reference Pritchard, Stephens and Donnelly2000) was used to identify clusters of individuals. This Bayesian approach, implemented in the software STRUCTURE, identifies the number of different subpopulations (K) and estimates the ancestry of the sampled individuals on the basis of their genotypes. We used a burn-in of 50,000 Markov Chain Monte Carlo (MCMC), a run length of 1,000,000 MCMC and a model allowing for admixture and correlated allele frequencies. Log-likelihood estimates were calculated for K=1 to 10 with six replicates each. The modal value of ΔK, a quantity based on the second order rate of change of the likelihood function with respect to K, was used to detect the number of clusters according to Evanno et al. (Reference Evanno, Regnaut and Goudet2005). Finally, the program DISTRUCT was used for graphical display of structure results (Rosenberg, Reference Rosenberg2002).
Isolation by distance was examined using the regression between F ST/(1−F ST) and the natural logarithm of distance (in kilometres) between sample populations. The null hypothesis of no geographical correlation to genetic divergence was examined by testing the significance of the rank correlation, using a nonparametric one-sided Mantel test based on 10,000 permutations. This was performed with the ISOLDE programme implemented in GENEPOP v. 4 (Rousset, Reference Rousset2007).
The effect of host plant and habitat (greenhouse versus open environment) was examined by performing an analysis of molecular variance (AMOVA) using Arlequin 3.0 (Excoffier et al., Reference Excoffier, Laval and Schneider2005) to estimate the different genetic variance components (among groups, among populations within groups, within populations).
Endosymbiont monitoring
To detect the presence of secondary symbionts, 16S rDNA for Rickettsia, Hamiltonella and Cardinium and 23S rDNA for Arsenophonus were amplified using genus-specific primers (Chiel et al., Reference Chiel, Gottlieb, Zchori-Fein, Mozes-Daube, Katzir, Inbar and Ghanim2007 and references therein). The presence of Wolbachia was determined using wsp primers (Zhou et al., Reference Zhou, Rousset and O'Neil1998). PCR conditions were as described in Chiel et al. (Reference Chiel, Gottlieb, Zchori-Fein, Mozes-Daube, Katzir, Inbar and Ghanim2007) and in Zhou et al. (Reference Zhou, Rousset and O'Neil1998). A total of 357 individuals from 19 populations (16–25 females per population with exception of NCr-23, 11 individuals) were examined by PCR for the presence of the above symbionts.
Wolbachia characterisation
Ten individuals randomly chosen from nine B. tabaci populations separated genetically and geographically were used for Multi Locus Sequence Typing (MLST) and wsp analysis (Baldo et al., Reference Baldo, Dunning Hotopp, Jolley, Bordenstein, Biber, Choudhury, Hayashi, Maiden, Tattelin and Werren2006). Amplification of the genes gatB, coxA, hcpA, fbpA, ftsZ and wsp and sequencing were performed using the universal primers described at http://www.pubmlst.org/Wolbachia/.
PCR amplifications were performed in 50 μl reactions containing 0.2 mM dNTPs, 1.5 mM MgCl2, each primer at a concentration of 1μM, Taq buffer (Minotech), 5 U Taq DNA polymerase (Minotech) and 2 μl DNA. Cycling conditions were: 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 1.5 min and a final elongation step at 72°C for 10 min. PCR products were purified using the Nucleospin Extract kit (Macherey-Nagel) and both strands directly sequenced (Macrogen, Korea). Editing of the sequences was performed using BioEdit v.7.0 (Hall, Reference Hall1999). Multiple sequence alignment was carried out using the software package Clustal W v.1.7 (Higgins et al., Reference Higgins, Thompson and Gibson1996). The sequences are deposited in the GenBank under the accession numbers JQ013512 and JQ013511, for Wolbachia ftsZ and coxA, respectively, JQ013509 and JQ013510 for Wolbachia gatB, JQ013507 and JQ013508 for Wolbachia fbpA, JQ013513 and JQ013514 for Wolbachia hcpA, and JQ013515 and JQ013516 for Wolbachia wsp.
Wolbachia typing
Polymorphisms were detected in genes gatB, hcpA, fbpA and wsp. Each gene had two allelic forms shared by five individuals, indicating the existence of two distinct Wolbachia strains. Sequences from our study, as well as sequences of Wolbachia from Bemisia in GenBank were aligned and subsequently examined for restriction recognition site with an in-house developed Perl script. For hcpA and gatB, no restriction sites suitable for routine discrimination between the two strains were found. Enzymes AluI and HhaI, respectively, allowed easy distinction between the two allelic forms of wsp and fbpA.
A 610 bp fragment of the wsp gene was amplified using the universal primers 81F and 691R (Zhou et al., Reference Zhou, Rousset and O'Neil1998) and subsequent digested with AluI. Digestion yields a restriction pattern with nine (27 bp, 23 bp, 24 bp, 48 bp, 50 bp, 54 bp, 84 bp, 93 bp and 207 bp) or ten (27 bp, 23 bp, 24 bp, 48 bp, 50 bp, 54 bp, 55 bp, 84 bp, 93 bp and 152 bp) fragments for the first and second allelic form, respectively. A 512 fragment of fbpA gene amplified using primers fbpA_F1 and fbpA_R1 (Baldo et al., Reference Baldo, Dunning Hotopp, Jolley, Bordenstein, Biber, Choudhury, Hayashi, Maiden, Tattelin and Werren2006) and subsequently digested with HhaI yields a restriction pattern with four fragments of 22 bp, 145 bp, 147 bp and 198 bp for the first allelic form and four fragments of 22 bp, 80 bp, 147 bp and 263 bp for the second allelic form.
PCR reactions were performed in 10 μl containing 0.2 mM dNTP, 0.2 μM primers, 0.5 U Taq DNA polymerase (DreamTaq, Fermentas) and 1 μl of DNA template. Cycling conditions were the same as for endosymbiont screening (see above). The PCR products were digested at 37°C for 3 h with 5 U of the respective enzyme (Fermentas Life Sciences), then incubated at 65°C for 20 min, and analyzed by electrophoresis on a 2% agarose gel.
The two PCR-RFLP based diagnostic assays were used to determine the prevalence of these Wolbachia strains in 19 Greek populations (approximately 12 individuals per population) representative of the geographic distribution of our sampling as well as of the different genetic populations.
The distributions of Wolbachia infection frequencies and the two Wolbachia strains between populations or groups of populations were compared with Fisher's exact test using the R statistical software (http://www.R-project.org). Mann-Whitney U-tests were also employed for some two by two comparisons.
Results
The sampling procedure does not allow discriminating males and females, so mixed sex individuals were collected from the field. However, only females were used in the genotyping and in all the molecular experiments including the monitoring of the symbionts. The selection of the female individuals was done under a binocular stereoscope based on the size and the shape of the posterior part of their bodies. This procedure revealed that in all of our samples both sexes were present.
Species identification
Among 38 Greek B. tabaci populations, the MEAM1 discriminative allele 268 of locus BT159 (called 273 in Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007) was found in 28 of 30 individuals from population NCr-216 collected on Lantana camara in a garden in north Crete. NCr-216 had also unique alleles for loci b34 and 145, whereas PCR primers for loci d26, 155 and 11 failed to amplify. The PCR-RFLP assay further demonstrated that population NCr-216 was a B population of MEAM1, and thus it was eliminated from further analysis. All other populations belonged to the Q B. tabaci. The PCR-RFLP assay, which discriminates between populations Q1 and Q2 (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007), was performed for 357 individuals of 19 populations, in which the bacterial community was also studied. All were of the Q1 B. tabaci. This confirms previous findings (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007; Roditakis et al., Reference Roditakis, Grispou, Morou, Kristoffersen, Roditakis, Nauen, Vontas and Tsagkarakou2009) that Q1 are the predominant B. tabaci in Greece.
Genetic polymorphism of the microsatellite loci
In all, 1134 females were genotyped at eight microsatellite loci. Heterozygote deficiency was especially high for locus BT-d26 in almost all populations (33/37). For this locus, analysis with Micro-Checker revealed the presence of null alleles. Thus, this marker was excluded from further analysis. Sixty-two individuals amplified for fewer than four loci and were removed from the analysis. The polymorphism of seven microsatellite markers for 1072 females belonging to 37 sampling populations were further analyzed. Mean number of alleles (N A), observed (H O) and expected (H E) heterozygosities as well as F IS values are given in table 1.
The number of alleles over all individuals ranged from four for locus 145 to 18 for locus BT-b34. The mean number of alleles per sample ranged from 2.57±0.79 to 4.71±1.81. The mean number of alleles was significantly lower in Crete (n=25, range from 2.57 to 4.71) than in mainland Greece, including the ISa and ISc samples (n=12, range from 3.42 to 4.71) as assessed by the Mann-Whitney test U=343, P<0.001.
H O values ranged from 0.24 to 0.45 and H E ranged from 0.285 to 0.506. In seven out of 37 populations, H O≥H E, resulting in negative F IS. Significant heterozygote deficits were found in 18 of the 37 populations. Most of the samples from mainland Greece (8/10) had significant heterozygote deficits. F IS values were also significantly lower in Crete (n=25) compared to mainland Greece (n=10) as assessed by the Mann- Whitney test U=235, P=0.042.
In order to test whether high F IS are associated with the sampling period, the relationship between significant heterozygote deficits and the colonization phase of the crop was investigated: samples collected during September–October for greenhouse crops and May–June for field crops were considered to be part of the early colonization phase. It was found that there were no significant differences (Fisher's exact test: P=0.191) between the samples collected early and after establishment on the crops.
Among the 720 tests performed for linkage disequilibrium, 25 were significant (in 17 populations) but none remained significant after the Bonferroni correction. Thus, the seven microsatellite loci carried independent genetic information.
Population structure
Bayesian clustering analysis of multilocus genotypes with STRUCTURE identified at least two clusters (K=2): lnP(D) increased from K=1 to K=4 and reached a plateau for K>4. The smallest value of K that captured the major structure in the data is 2. Most individuals (1005/1072) are clearly assigned to one of the two clusters (Q>90) (fig. 2). The first cluster contains only the individuals from mainland Greece, the islands of Santorini and Schoinoussa and north Crete. The second genetic cluster contains only samples from south Crete. In terms of allelic frequencies, the most striking difference between the two clusters concerns locus Bt-155. Allele 198 of this locus has in the first cluster a frequency of 0.001 and in the second cluster a frequency of 0.708, while allele 196 is found with a frequency of 0.001 in the second and with a frequency of 0.644 in the first cluster. Only a few individuals (67/1072) had admixed ancestry.

Fig. 2. Results of STRUCTURE clustering analysis (K=2) of 1072 B. tabaci belonging to 37 sampling populations. Individuals are presented by vertical lines, divided into segments that represent their inferred membership into each of the two clusters.
Gene flow in relation to geographic distribution
Genetic differentiation was analyzed by relating genotypic distribution with F ST estimates. Differentiation was highly significant (P<10−5) over all samples (F ST=0.15) (table 3), as well as among samples collected in Crete (F ST=0.13). The same was true for samples from south Crete F ST=0.1, P<10−5. Significant differentiation (P=0.04) was also found for samples collected in mainland Greece; however, there was more gene flow than among Cretan samples with lower F ST (F ST=0.0046). Among the ten samples from mainland Greece, only MGr-69 was significantly different from samples MGr-76, 92 and 94 (F ST=0.014, 0.018, 0.017; P=0.012, 0.044, 0.012, respectively). All other samples from mainland Greece were not differentiated from each other, with pairwise F ST ranging from −0.0001 to 0.018 and P>0.05, even where geographic distances exceed 500 km. For the Cretan samples, however, significant differentiation (P<0.05) was identified in most of the pairwise comparisons (260/300). The highest level of differentiation among the Crete populations, and at the same time among all pairwise comparisons in this study, was found between samples SCr-190 and SCr-29 (F ST=0.331, P<0.001).
Table 3. F ST estimates for population subdivisions of B. tabaci at different geographic scales.
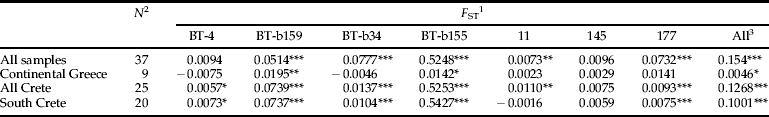
1 Estimated by the estimator of Weir & Cockerham (Reference Weir and Cockerham1984).
2 Number of samples considered.
3 “All” refers to the multi-locus estimate.
Significant genotype differentiation: *P<0.05, **P<0.01, ***P<0.001.
Isolation by distance
Mantel tests were performed to assess isolation by distance. The relationship between gene flow and geographic distance was analyzed by examining the slope of the regression function between multilocus pairwise estimates of F ST/(1−F ST) and log (geographic distance), computed for each pair of samples. We analyzed (i) all sampled populations of B. tabaci, (ii) all populations from Crete, (iii) all populations from mainland Greece and (iv) all populations from south Crete. For all populations, the slope of the regression was positive (b=0.024) with a significant positive rank correlation between F ST/(1−F ST) and (ln) geographic distance (Mantel test, P<0.001). The significant positive correlation remains (b=0.01281, P<0.003) for the samples from mainland Greece and north Crete, which clustered together in the same genetic group (see above). However, when considering only samples from mainland Greece (n=10) or only samples from Crete (n=23) there is no significant correlation: b=−0.0001, P=0.34 and b=0.0094, P=0.13 for mainland Greece and Crete, respectively. These results indicate that gene flow between populations decreases with geographic distance but that, at a smaller geographic scale, factors other than geographic isolation dominate in shaping the genetic structure.
The role of host plant species and habitat in B. tabaci differentiation
When considering all populations, AMOVA analysis showed that the variance within populations explained 84.72% of the total variance, while the rest was due to variance among populations (table 4). Considering the two genetic groups, AMOVA showed that 16.62% of the variance was attributed to differences between these two clusters and 5.52% to differences among populations in the two clusters.
Table 4. Analysis of molecular Variance (AMOVA) for B. tabaci using seven microsatellite loci (1) among all populations, (2) between the two groups identified by STRUCTURE, (3) among three groups of plants (see text for details) and (4) between greenhouses versus field samples (only south Cretan samples are considered).

Host plant adaptation should restrict gene flow between populations colonizing different plant species. Similarly, adaptation to specific conditions in greenhouses should restrict gene flow between populations inhabiting greenhouses and populations in the open environment. To determine the impact of the host plant on the genetic affinities of the B. tabaci samples, an analysis based on clusters of the three representative families of host plants sampled in this study was performed. When considering the three groups with respect to the three host plant families from which B. tabaci was mainly sampled (Solanaceae: 16 samples, Curcubitacea: 12 samples and Malvacea: four samples) or to the habitat type (only for the Cretan populations, 12 samples from greenhouses versus 11 samples from open environment), the genetic variability among groups accounted only for the 0.48% (host plant) and −0.18% (greenhouse/open environment) of the global genetic diversity and the fixation index among groups (Fct) was not significant (Fct=0.0047, P=0.258; and Fct=−0.00181, P=0.44), suggesting that populations were still structured within at least one group. This indicates that neither host plant species nor the habitat type play a role in the genetic differentiation in our samples.
The results of the AMOVA confirmed the presence of structure in our samples, which is not associated with the host plants or the habitat type.
Presence of secondary symbionts
The presence of secondary symbionts in 309 individuals from 19 populations was examined. All individuals were associated with Hamiltonella, while there was no evidence for the presence of Arsenophonus, Rickettsia or Cardinium. The three reactions failed to produce amplification products with the test insects but produced successful amplification with known positive controls (Q2 from Israel for Arsenophonus, and Rickettsia and Q1 from Spain for Cardinium). We partially sequenced the PCR products of the 16S ribosomal RNA gene of Hamiltonella from six B. tabaci individuals from different localities, finding no polymorphism among individuals. The sequences were identical to other Hamiltonella sequences already published.
The prevalence of Wolbachia varied among populations from 45% to 100% (average: 75%; fig. 3). The variation in Wolbachia prevalence was significant when all the 19 populations were considered separately (Fisher's Exact test, P<0.0005) but also when populations from the same region (SCr, NCr, MGr) were grouped together (P<0.0005). Wolbachia prevalence is lower in mainland Greece (average 59%) than in south (84%) or north (91%) Crete (P<0.0001). The difference between south and north Crete is not significant (P=0.47). Within regions, the highest variability of Wolbachia prevalence among populations is found in south Crete (P=0.008), while differences are not significant among populations in north Crete or in mainland Greece (P>0.08).

Fig. 3. Frequency of Wolbachia negative and of Wolbachia strain W1 and Wolbachia strain W2 in 19 populations of B. tabaci from Greece. Populations are ranked with increasing frequency of W1. The number of individuals tested for the presence of Wolbachia is given in brackets after the name of the population. Significant heterozygote deficits are given before the name of each population: *P<0.05, **P<0.01, ***P<0.001; ![]() , not infected;
, not infected; ![]() , W2;
, W2; ![]() , W1.
, W1.
Wolbachia characterisation
Ten individuals randomly chosen from nine B. tabaci populations separated genetically and geographically were used for MLST and wsp analysis (Baldo et al., Reference Baldo, Dunning Hotopp, Jolley, Bordenstein, Biber, Choudhury, Hayashi, Maiden, Tattelin and Werren2006).
Among the six genes tested, no polymorphism was detected for coxA and ftsZ, while the others (gatB, hcpA, fbpA, wsp) showed two allelic forms, each shared by five individuals, indicating the presence of two distinct Wolbachia strains (table 5). However, we can't exclude that other strains are present in these populations. Indeed, as PCR products were directly sequenced, in case of multiple infections, a strain in very low proportion may not be detected.
Table 5. Results of the MLST analysis as disclosed by the partial sequences of five genes obtained for ten B. tabaci individuals.
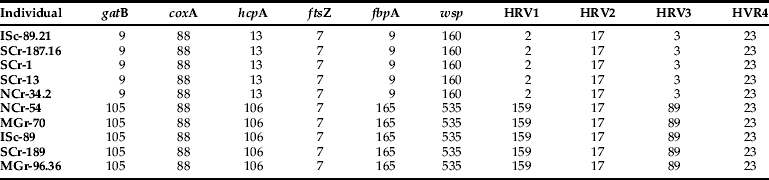
Comparison against the multilocus sequence typing (MLST) database (http://pubmlst.org/Wolbachia/) and the wsp database showed that strain W1 carries alleles 9, 88, 13, 7, 9 and 160 of loci gatB, coxA, hcpA, ftsZ, fbpA and wsp, respectively. The second stain (W2) carries alleles 105, 88, 106, 7, 165 and 535 of loci gatB, coxA, hcpA, ftsZ, fbpA and wsp, respectively. Phylogenetic analysis based on the ftsZ sequences indicated that the strains belong to the Wolbachia group B, as previously described Wolbachia strains found in B. tabaci (Nirgianaki et al., Reference Nirgianaki, Banks, Frohlich, Veneti, Braig, Miller, Bedford, Markham, Savakis and Bourtzis2003).
Typing of Wolbachia
Digestion with AluI of a 610 bp fragment of the wsp gene yielded two distinct restriction patterns discriminative for W1 and W2, respectively (fig. 4). Similarly, digestion with HhaI of a 512 bp fragment of the fbpA gene yielded two distinct restriction patterns discriminative for W1 and W2 (see fig. 4).

Fig. 4. PCR-RFLP assays to discriminate between the two Wolbachia strains. (A) Couple marker-enzyme wsp/AluI. 1, 100 bp Ladder; 2, 3, strain W1; 4, strain W2. The strains W1 and W2 can be distinguished by a band at 207 and 156 bp, respectively. (B) Couple marker-enzyme fbpA/HhaI. 1, 100 bp ladder; 2, 3, strain W1; 4, strain W2. The strains W1 and W2 can be distinguished by a band at 263 and 198 bp, respectively. Note that all other fragments <60 bp expected after digestion (22 bp, 23 bp, 24 bp, 27 bp, 48 bp, 50 bp, 54 bp and 55 bp) were not detected as they were too small to be visualized by electrophoresis in a routine agarose gel.
This PCR-RFLP diagnostic test was applied to 233 infected individuals from 19 populations to determine Wolbachia infections. For half of the individuals (randomly chosen), PCR-RFLP were done with the two marker-enzyme pairs. Since we always found the same wsp and fbpA alleles to be coupled, the other half of samples was only tested with fbpA/HhaI.
We did not detect double infections in any of the individuals. However, we cannot exclude double infections in which one of the co-infecting strains is strongly underrepresented.
The two strains did not occur with equal frequency: W2 was present in 68% of the Wolbachia-infected B. tabaci individuals. This varied from 6% to 100% in different populations (Fisher's Exact test, P<0.0005) and was significant different between MGr and SCr or NCr (P<0.0005). The highest frequency of W2 was found in mainland Greece (90.5%). In south and north Crete the frequencies were 50.3% and 59.9%, respectively, but not significantly different (P=0.38). Within each of the three regions, the differences are significant only in SCr (P=0.002), P>0.09 for MGr and NCr. In Crete, the strain W1 was found in higher frequency in samples collected in 2002 (table 1 and fig. 3), but its presence decreased in samples from subsequent years.
Infection patterns in relation to the population structure
There was no significant correlation between either F IS and the frequency of Wolbachia-infected whiteflies (R=0.148, P=0.5456) or F IS and the prevalence of the one or the other Wolbachia strains (R=−0.2945 and R=0.2412 for strains W1 and W2, respectively; P>0.05). When the distributions of Wolbachia infection and Wolbachia strains are compared between populations with significant HW departures and populations that are under HW equilibrium, no differences in the frequency of infected individuals were found (mean infection frequency in populations with significant heterozygote deficits=66.19±16.59, n=10, and in populations under HW equilibrium=81.63±19.02, n=9; U=99, P=0.22); however, a significantly lower prevalence of Wolbachia strain W1 is found in the populations with heterozygote deficits (mean frequency of W1 in populations with significant heterozygote deficits=15.15±13.08, n=10, and in populations under HW equilibrium=37.71±27.45, n=9; Fisher's Exact test, P<0.0005).
The two different genetic groups (populations SCr versus populations MGr/NCr) have distinct infection patterns: (i) in the first group there is a higher frequency of Wolbachia-infected individuals compared to the second genetic group (Fisher's Exact test, P=0.003), with mean infection in the MGr/NCr group of 66.04±18.57 and mean infection in the SCr group of 83.77±15.09; and (ii) the two genetic groups differ in relation to the Wolbachia strain prevalence (Fisher's Exact test, P<0.0005). The latter is due to the lower frequency of Wolbachia strain W1 in the MGr/NCr samples (mean frequency of infected individuals with strain W1 per population is 14.08±17.34, n=11) compared to the south Cretan samples (mean frequency of infected individuals with strain W1 per population=42.00±22.96, n=8).
However, when looking at pairwise comparisons of populations, there is no clear influence of Wolbachia on gene flow. For example, population SCr-195, infected mostly with the W2 strain (fig. 3), is genetically closer to SCr-1, which is mostly infected with W1 (F ST=0.036, P=0.0043) than to population SCr-191 with a similar infection pattern (F ST=0.2212, P<0.001).
Discussion
The genetic structure of B. tabaci from mainland and insular Greece was studied by investigating polymorphic microsatellite loci. Thirty-seven out of 38 populations were assigned to the Q B. tabaci, which is predominant in this geographic area (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007; Roditakis et al., Reference Roditakis, Grispou, Morou, Kristoffersen, Roditakis, Nauen, Vontas and Tsagkarakou2009). All Q individuals examined belonged to the west Mediterranean Q1 populations (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007). Strong genetic differentiation was found among the sampled populations of the whole data set. A Bayesian approach disclosed that the individuals clustered into at least two groups (G1 and G2) based on their genotypes. Two genetic populations in Greek samples were also found in a previous exploratory work (Tsagkarakou et al., Reference Tsagkarakou, Tsigenopoulos, Gorman, Lagnel and Bedford2007); however, due to the limited number of samples, no correlation to the geographic origin of the samples was possible. In the present study, we found that B. tabaci were highly differentiated and displayed a strong geographic structure: those from south Crete all formed one genetic group (G1), while those from north Crete were in the same cluster (G2) together with samples from mainland Greece and from the islands Schoinoussa and Santorini. Within G1, the B. tabaci populations were highly differentiated. Populations within G2 were also differentiated; however, the differentiation within the populations from mainland Greece was the lowest observed, even when separated by more than 100 km. These results show that B. tabaci populations from Greece are genetically structured.
Host plant
The formation of sympatric differentiated populations as a consequence of host plant adaptation to plant species or even at the finer level of individual plant genotypes has been suggested for phytophagous insects (Mopper, Reference Mopper1996). B. tabaci is one of the few whitefly species associated with herbaceous hosts with an ability to attack multiple crop, weed and ornamental hosts. The different performance on diverse host species has been attributed to different biotypes (Maruthi et al., Reference Maruthi, Colvin, Thwaites, Banks, Gibson and Seal2004) and might be in the origin of the genetic differentiation found among B. tabaci populations (De Barro, Reference De Barro2005). In our study, considering the family level, the analysis of molecular variance indicates that the differentiation does not originate in the different host plants colonized. Gene flow was not higher between populations collected on species belonging to the same plant family compared to gene flow between populations from different plant families.
Habitat
Populations from south Crete were sampled from greenhouses and plants in the open environment. There was significant differentiation among them, even between populations separated by only a few meters (e.g. pop SCr189 and SCr190 (F ST=0.035). However, the genetic differences were not explained by the different habitats. The analysis of molecular variance showed no correlation of the type of habitat with the molecular differences between samples from southern Crete. Populations from greenhouses were not genetically closer among them than to B. tabaci from outside greenhouses. Due to the mild weather (minimum temperatures are often above 10°C in winter), populations of B. tabaci can be found in southern Crete all year, infesting vegetable crops and weeds. During the six colder months (November–April), when cultivation occurs mostly in greenhouses, B. tabaci invades protected crops but can also be found outside greenhouses on weed hosts. During the six summer months, it infests the outdoor cultivations. This continuous cycle does not permit a homogeneous ‘greenhouse’ or ‘outdoor’ population to be formed. On the other hand, the repetitive colonisations promote genetic differences due to genetic drift. This phenomenon is accentuated in south Crete due to the intensive use of insecticide treatments, which induces large fluctuations of population sizes.
Intense selection through extensive Bacillus thuringiensis applications used in some greenhouses significantly impacted the genetic structure of Trichoplusia ni populations in Canada and resulted in a strong positive correlation between Bt resistance, and genetic differentiation (Franklin et al., Reference Franklin, Ritland and Myers2010). A similar relationship was found in the French codling moth, Cydia pomonella, where the mean number of insecticide sprays was positively correlated with levels of genetic differentiation (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007). In southern Crete, where B. tabaci control relies on intensive insecticide use and high resistance levels are found (Roditakis et al., Reference Roditakis, Grispou, Morou, Kristoffersen, Roditakis, Nauen, Vontas and Tsagkarakou2009), the frequency of the different resistant alleles varied, underlying the presence of differentiated populations (Tsagkarakou et al., Reference Roditakis, Grispou, Morou, Kristoffersen, Roditakis, Nauen, Vontas and Tsagkarakou2009).
Geographic distance
Significant isolation by distance is found when the whole data set is considered; increase of geographic distance decreases gene flow between populations. However, populations from northern Crete are genetically closer to those from mainland Greece than those from southern Crete. Also, the absence of isolation by distance when considering only samples from mainland or only samples from Crete indicate that factors other than the geographic distance play a major role in the genetic structure of B. tabaci in Greece. The landscape fragmentation due to the mountain range in the middle of Crete may contribute to the differences between samples from northern Crete and southern Crete. Landscape fragmentation does not play a significant role in mainland Greece since samples collected there formed almost a panmictic population (except one sample). The absence of high differentiation in mainland Greece is in accordance with the findings of Dalmon et al. (Reference Dalmon, Halkett, Granier, Delatte and Peterschmitt2008) for greenhouse B. tabaci in France where recent colonization of the species, together with human activities, have been evoked to explain the high F ST among samples. However, conditions are different for B. tabaci in mainland Greece where the species has been established at least since 1889, when it was first described by Gennadius (Reference Gennadius1889). The homogenization observed in mainland Greece may be due to the fact that B. tabaci inhabits mainly outdoor cultivations and not greenhouses, where Trialeurodes vaporariorum remains the main whitefly pest. Also, members of the B. tabaci complex can disperse over long distances despite its small size. Dispersal is not entirely passive and may not be considered weak since it is capable of sustaining flight in the field for distances of up 7.0 km in a 12-h period (Byrne, Reference Byrne1999).
Before 2000, B. tabaci was not considered an important pest in greenhouses in Crete (Roditakis N, personal communication). Since then, it has replaced T. vaporariorum (at least in south Crete) and is currently considered the major pest in combination with virus transmission outbreaks. One hypothesis explaining this situation is that a particular genetic population of B. tabaci has been installed in southern Crete, which does not move much since suitable host plants and climate conditions are present year round. Under these conditions, southern Cretan populations of B. tabaci would rarely move more than a few meters although their dispersal capacities are higher. This, together with the intensive bottlenecks imposed by the repetitive heavy pesticide applications in the confined environment of greenhouses could explain the absence of isolation by distance in Crete, as well as the high differentiation among samples due to genetic drift.
Secondary symbionts
Insects have established symbiotic associations with diverse groups of bacterial species. These associations affect various aspects of insect host biology, including development, nutrition, reproduction, speciation, immunity, vector competence, as well as host preference (Moran et al., Reference Moran, McCutcheon and Nakabachi2008). We tested for the presence of Hamiltonella, Rickettsia, Wolbachia, Cardinium and Arsenophonus in Greek populations of Q1 B. tabaci. Only Hamiltonella and Wolbachia were present, while Rickettsia, Cardinium and Arsenophonus were absent from all individuals tested. Hamiltonella infection is fixed in all populations, while only a patchy distribution of Wolbachia was observed.
The role of Hamiltonella in the Greek populations of B. tabaci is currently unknown. In aphids, the presence of Hamiltonella (and its APSE phage) provides protection against natural enemies (Oliver et al., Reference Oliver, Degnan, Burke and Moran2010); however, in the present study, we did not examine whether the Hamiltonella of the Greek B. tabaci is infected by the APSE phage. Gottlieb et al. (Reference Gottlieb, Ghanim, Gueguen, Kontsedalov, Vavre, Fleury and Zchori-Fein2008) reported that Hamiltonella was located within the bacteriocytes of B. tabaci and MEAM1 together with the primary symbiont Portiera. Whether Hamiltonella is acting as a co-primary symbiont or is competing with Portiera is still under question (Gottlieb et al., Reference Gottlieb, Ghanim, Gueguen, Kontsedalov, Vavre, Fleury and Zchori-Fein2008). Alternatively, these two endosymbionts may have a complementary metabolic role, like Buchnera aphidicola and Serratia symbiotica in tryptophan provision in the aphid Cinara cedri (Gómez-Valero et al., Reference Gómez-Valero, Soriano-Navarro, Pérez-Brocal, Heddi, Moya, García-Verdugo and Latorre2004).
It is also worth noting that the presence of Hamiltonella and the expression of its GroEL protein were recently correlated with the transmission efficiency of Tomato Yellow Leaf Curl Virus (TYLCV) by MEAM1 (Gottlieb et al., Reference Gottlieb, Zchori-Fein, Mozes-Daube, Kontsedalov, Skaljac, Brumin, Sobol, Czosnek, Vavre, Fleury and Ghanim2010). Furthermore, a mutualistic relationship between plant virus and the insect host has been shown in MEAM1 from China, which increased their fecundity, longevity and population density when feeding on Begomovirus-infected plants (Jiu et al., Reference Jiu, Zhou, Tong, Xu, Yang, Wan and Liu2007). If such an impact on host phenotype applies in the case of TYLCV and members of the B. tabaci complex, this fitness benefit could indirectly explain the high prevalence of Hamiltonella associated with TYLCV transmission efficacy. This hypothesis cannot explain on its own the 100% fixation of Hamiltonella in the present study. Jiu et al. (Reference Jiu, Zhou, Tong, Xu, Yang, Wan and Liu2007) showed that the mutually beneficial relationships between the invasive whitefly and the plant viruses are indirect via the plants; however, our study included also B. tabaci from plants which are not infected by TYLCV.
It is known that the presence of Wolbachia in insects is usually involved in the manipulation of the host's reproductive system, including feminization, parthenogenesis, male killing and cytoplasmic incompatibility (Saridaki & Bourtzis, Reference Saridaki and Bourtzis2010). Wolbachia was detected in Greek populations of B. tabaci. Although the exact sex ratio was not determined in each population studied here, it is worth mentioning that both sexes were present in the tested B. tabaci. The prevalence of Wolbachia varied between populations and localities, with the lowest abundance in mainland Greece. Previous studies have reported the presence of Wolbachia, as a single infection, in natural populations of the B. tabaci complex (Nirgianaki et al., Reference Nirgianaki, Banks, Frohlich, Veneti, Braig, Miller, Bedford, Markham, Savakis and Bourtzis2003; Li et al., Reference Li, Lin and Guo2007). Gueguen et al. (Reference Gueguen, Vavre, Gnankine, Peterschmitt, Charif, Chiel, Gottlieb, Ghanim, Zchori-Fein and Fleury2010) recently reported the presence of the so-called W1 Wolbachia strain in Q1 populations, while the W2 Wolbachia strain is associated with the Q2 from Israel. However, no individual of B. tabaci has ever been reported to be double- or multiple-infected. We report for the first time the presence of two Wolbachia strains (W1 and W2) in the same Q1, at the population but not at the individual level. The apparent lack of double infections could be due to: (i) the presence of a Wolbachia strain at a low, PCR non-detectable infection level. Such low-titer Wolbachia strains have been recently described in different insect host species (Arthofer et al., Reference Arthofer, Riegler, Schneider, Krammer, Miller and Stauffer2009) and (ii) the recent invasion of a novel Wolbachia strain into previously infected natural populations. Indeed, our data suggest that hypothesis (ii) may be valid. Although, in general, W2 was the most frequent strain in infested individuals from mainland Greece and W1 the most frequent strain in infested individuals from south Crete, there is no clear association of the two Wolbachia strains (W1 and W2) with the two genetic groups (SCr and MGr/NCr). However, two findings suggest a possible role of the W2 Wolbachia strain in the genetic structure of the Greek populations of B. tabaci Q1: (i) Wolbachia strain W1 is present at a low frequency in populations with significant heterozygote deficiency (MGr/NCr genetic group) compared to the Hardy-Weinberg equilibrium populations (SCr genetic group) and (ii) Wolbachia strain W2 is more prevalent than strain W1 in all populations studied, except in four populations (SCr-23, 13, 1 and NCr-17) from southern and northern Crete. It is important to note here that these four samples are the oldest included in our survey and were originally collected in 2002. If the Wolbachia strain W2 is incompatible with strain W1 (the ‘ancient’ strain), then our data are consistent with the scenario of a replacement of W1 by W2 in Greek populations of Q1 B. tabaci. Compatibility assays, as well as monitoring of the Wolbachia strains W1 and W2 in a B. tabaci collection over time, are both required in order to evaluate the CI properties of these strains.
It was recently shown that Wolbachia is present both inside and outside bacteriocytes of Israeli populations of Q B. tabaci, presumably Q2, which is known to harbor the Wolbachia strain W2 (Gottlieb et al., Reference Gottlieb, Ghanim, Gueguen, Kontsedalov, Vavre, Fleury and Zchori-Fein2008; Gueguen et al., Reference Gueguen, Vavre, Gnankine, Peterschmitt, Charif, Chiel, Gottlieb, Ghanim, Zchori-Fein and Fleury2010). Another recent study showed that Wolbachia is confined inside bacteriocytes in Croatian populations of Q B. tabaci (Skaljac et al., Reference Skaljac, Zanic, Goreta Ban, Kontsedalov and Ghanim2010). It is known that both Q1 and Q2 B. tabaci exist in Croatia (Dinsdale et al., Reference Dinsdale, Cook, Riginos, Buckley and De Barro2010); however, neither the whitefly populations nor the Wolbachia strain(s) present in them were genotyped (Skaljac et al., Reference Skaljac, Zanic, Goreta Ban, Kontsedalov and Ghanim2010) to know if the outside- and/or inside-bacteriocyte localization pattern is a specific genetic property of a particular Wolbachia strain or symbiotic association. This question, as well as the mode and efficiency of transmission of W1 and W2 Wolbachia strains, are currently under investigation in our laboratories. Given the fact that recent reports suggest dynamic interactions of Wolbachia with different viruses (Bian et al., Reference Bian, Xu, Lu, Xie and Xi2010) the role of both W1 and W2 Wolbachia strains in the transmission of plant viruses by Q B. tabaci also requires a thorough analysis.
In conclusion, we show that among Q1 B. tabaci gene flow is often very restricted. Bayesian analysis suggests that whiteflies from Greece group in two genetic populations; geographic and anthropogenic parameters seem to play a major role in this grouping. The high prevalence of Hamiltonella and Wolbachia may not be related with the occurrence of these two genetic groups; however, the role of the Wolbachia strains W1 and W2 in the shaping of the population genetic structure has to be examined in a broader B. tabaci sampling and by performing reproductive compatibility assays.
Acknowledgements
We thank Hélène Henri for her valuable contribution in performing the PCR-RFLP diagnostic assay and Dr Antonis Magoulas for giving us access to sequencing facilities in the Marine Biology and Genetics Institute, HCMR, Heraklion. This work was partially supported by a General Secretariat of Research and Technology bilateral Greece-France (229-E) to AT and by EU CSA-REGPROT 203590 – MicrobeGR and intramural funds of the University of Ioannina to KB. KB also benefited by travel grants from EU Cost Action FA0701: ‘Arthropod Symbioses: From Fundamental Studies to Pest and Disease Management’.











