Introduction
In order to protect ecosystems, it is necessary to measure to what extent they have been altered. A widely adopted approach for evaluating aquatic ecosystems has been analysis of the benthic macroinvertebrate (BMI) community (Carew et al., Reference Carew, Pettigrove and Hoffmann2003). The larvae of Chironomidae, commonly called chironomids or bloodworms, are usually a major component of the BMI community. They are abundant and distributed globally, live in almost any aquatic habitat, tolerate a wide range of salinities and form an integral part of the diet for both vertebrate and invertebrate organisms – placing them in an important position in the aquatic foodweb. Therefore, chironomids have been frequently used for assessment of both acute and sublethal toxicity of contaminated sediments and water (Epler, Reference Epler2001). Chironomids are holometabolous insects that pass through four distinct stages in their life cycle. They spend the greatest part of their life cycle in larval form (Ebrahimnezhad & Fakhri, Reference Ebrahimnezhad and Fakhri2005; Chetelat et al., Reference Chetelat, Amyot, Cloutier and Poulain2008). Worldwide, there may be more than 10,000 species of chironomids (Armitage et al., Reference Armitage, Cranston and Pinder1995). Their distribution is closely related to dissolved oxygen (DO), organic matter, temperature, and different degrees of water depth – different species of larvae can live in/on sediments and vegetation (Das & Handique, Reference Das and Handique1996; Epler, Reference Epler2001; Ebrahimnezhad & Fakhri, Reference Ebrahimnezhad and Fakhri2005). Methods for culturing and testing are established by the US Environmental Protection Agency (US EPA), which has a standard protocol for testing toxic sediments using chironomids (US EPA, 1996).
Determining the species of chironomids is important since there is substantial variation among species in their response to environmental change; species-level identification provides the most relevant data when using population and community-level parameters to evaluate a study site (Carew et al., Reference Carew, Pettigrove and Hoffmann2003). Determining the correct genus and species can be time consuming – involving processing of BMI samples and making slide mounts. Head capsule morphology of chironomid larvae has been a principle means for taxonomically identifying larval species (Cranston, Reference Cranston2000; Epler, Reference Epler2001). It can require a high level of taxonomic skill. Large-scale routines often lead to misidentifications even when identification is restricted to genera, families or orders (Epler, Reference Epler2001; Carew et al., Reference Carew, Pettigrove and Hoffmann2003; Pfenninger et al., Reference Pfenninger, Nowak, Kley, Steinke and Streit2007). A source of error when using head capsule morphology for taxonomic identification is head capsule deformity. Head capsule deformities of chironomid larvae can be a common biomarker for monitoring environment health.
Morphological abnormalities such as antennal and mouthpart deformities are dependable indicators of various anthropogenic stressors in aquatic systems and in assessments of ecological risk (Warwick, Reference Warwick1985; Madden et al., Reference Madden, Suter, Nicholson and Austin1992; Swansburg et al., Reference Swansburg, Fairchild, Fryer and Ciborowski2002; Martinez et al., Reference Martinez, Moore, Schaumloffel and Dasgupta2003). In addition, deformities in mouth parts can result from breakage incurred when mounting head capsules. Therefore, one goal of this study was to develop an alternative method for taxonomic identification of wild chironomids that would facilitate their use in field work.
Molecular approaches have emerged over recent years to assist with this problem. Approaches have included various deoxyribonucleic acid (DNA) markers such as two ribosomal genes (18S and 28S), a single mitochondrial protein-coding gene (cytochrome oxidase I) and one nuclear protein-coding gene (CAD). These DNA markers have been generated using polymerase chain reaction (PCR)-based techniques including PCR-restriction fragment length polymorphism (RFLP), nucleotide sequence alignment, and DNA barcoding (Guryev et al., Reference Guryev, Makarevitch, Blinov and Martin2001; Carew et al., Reference Carew, Pettigrove and Hoffmann2003; Pfenninger et al., Reference Pfenninger, Nowak, Kley, Steinke and Streit2007; Cranston et al., Reference Cranston, Hardy and Morse2012). There are advantages to PCR-based assays. They are widely available, relatively inexpensive, require only small amounts of DNA and can be used to amplify gene-specific targets. However, even these PCR-based strategies raise concerns such as minimal start-up information for some species, resulting in considerable effort for generating and optimizing primers, and limited ability to screen polymorphic sites in the target DNA region resulting in non-specific genotyping. After all, the major challenge remains as these PCR-based approaches require an extraction of high-quality DNA from large numbers of samples – leading to an increase in processing time and error rates – and therefore, may not be the most accurate and cost-effective approaches (Pfrender et al., Reference Pfrender, Hawkins, Bagley, Courtney, Creutzburg, Epler, Fend, Ferrington, Hartzell, Jackson, Larsen, Levesque, Morse, Petersen, Ruiter, Schindel and Whiting2010).
Hemoglobin (Hb) proteins may offer an alternative to PCR techniques. Hb proteins in invertebrates are complex and have different overall architectures. In chironomids, Hb proteins are synthesized by the insect's fat body and secreted directly into hemolymph starting in the second instar (Bergtrom et al., Reference Bergtrom, Laufer and Rogers1976). Concentrations of Hb proteins gradually increase as the larvae reach later instars (Schin et al., Reference Schin, Poluhowich, Gamo and Laufer1974; Bergtrom et al., Reference Bergtrom, Laufer and Rogers1976). Due to high concentrations of Hb in hemolymph (Tichy Reference Tichy1975), large amounts are easily obtained from individuals for research purposes (Bentivegna et al., Reference Bentivegna, Oh, Doan and DiPietro2009). The abundance and presence of Hb in chironomids is physiologically relevant. They allow the larvae to sustain aerobic metabolism under adverse environment conditions such as polluted and hypoxic sediments (Lee et al., Reference Lee, Lee, Park and Choi2006). Hb proteins in the genus Chironomus show a high degree of polymorphism in addition to a high affinity for oxygen (Osmulski & Leyko, Reference Osmulski and Leyko1986). Hb polymorphism is stage-, species-, and tissue-specific, where different species have different numbers of Hbs. For example, Chironomus thummi has as many as 10 and Chironomus tentans has up to 14 Hb proteins (Bergtrom et al., Reference Bergtrom, Laufer and Rogers1976; Vafopoulou-Mandalos & Laufer, Reference Vafopoulou-Mandalos and Laufer1982; Schmidt et al., Reference Schmidt, Keyl and Hankeln1988). The architecture of the Hb molecule includes one (monomer) or two subunits (dimer) per molecule instead of the four typical of vertebrates (Wollmer et al., Reference Wollmer, Buse, Sick and Gersonde1972; Osmulski & Leyko et al., Reference Osmulski and Leyko1986; Das & Handique, Reference Das and Handique1996). The heterogeneity characterizing chironomid Hbs may be adaptive to exogenous and endogenous factors. This is demonstrated by the markedly higher contribution of dimeric Hbs in hemolymph of summer larvae compared with spring larvae of C. thummi thummi (CTT) (Osmulski & Leyko, Reference Osmulski and Leyko1986).
The purpose of this study was to develop Hb protein polymorphism as a tool for chironomid identification to support and supplement existing taxonomic keys, which are morphology- or PCR-based. Morphology-based keys typically identify larval chironomids using their head capsules and posterior appendages. Approaches for chironomid identification are needed as the most field studies utilizing BMI collect the larval stage of this family, which may not be as readily identifiable as the adult stage. The objective of the work presented here was to determine whether or not sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) would allow chironomid species to be identified by the size and intensity of their different Hb bands. Previous work in our laboratory showed that chironomid Hb proteins separated by SDS–PAGE can be detected using only one individual and Commaisse Blue staining (Bentivegna et al., Reference Bentivegna, Oh, Doan and DiPietro2009). The presence, absence and intensity of bands could be seen with the naked eye. This technique resulted in a significant reduction in labor and cost as no antibodies were needed. It also allowed the Hb pattern from SDS–PAGE to be compared to the head capsule and body of the same individual so that Hb proteins and morphology could be linked.
Materials and methods
Description of sampling sites
Chironomids were collected from four different sampling sites, comprised of one river and three wetlands, all located on the east coast of the USA during summers of 2007, 2008, and 2012. Two sites were from the state of New Jersey and the other two sites were from the state of Maine (table 1). Kearny Marsh (KM) of the New Jersey Meadowlands is located between the Passaic and Hackensack Rivers and is surrounded by major highways such as Interstate 95. High levels of toxic heavy metals have been found in this marsh (Bentivegna et al., Reference Bentivegna, Alfano, Bugel and Czechowicz2004). Rahway River (RR) is located in a residential area in Essex County of New Jersey. Chironomids were collected from the east branch which runs through the town of South Orange. North East Creek (NEC) and Bass Harbor (BH) are located in Mount Desert Island, Maine, with close proximity to sea water. Both sites are near Acadia National Park, which is considered to be a relatively pristine area when compared to the New Jersey watershed (US EPA, 2010a, b).
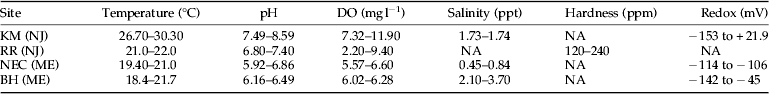
Table 1. Range of water quality parameters at the four chironomid collection sites.
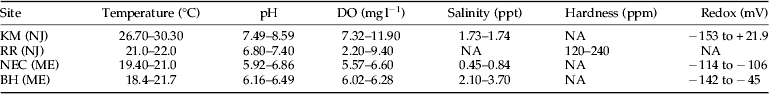
DO, dissolved oxygen; Redox, reduction and oxidation potential; NA, not available; KM (NJ), Kearny Marsh, New Jersey; RR (NJ), Rahway River, New Jersey; NEC (ME), North East Creek, Maine; BH (ME), Bass Harbor, Maine.
Chironomid collection
Chironomids in Maine were collected using a Hester–Dendy sampler (Wildco, Yulee, FL) or by hand picking them from rocks and aquatic vegetation. Chironomids in KM were collected using Hester–Dendy, while those in the RR were collected using Surber samplers (Wildco). The live larvae were placed into a Ziploc bag containing site water and transported back to the laboratory in a cooler. One to four hours transpired between chironomid collection and Hb protein collection (see below). In addition to the wild larvae, investigations included a laboratory strain of Chironomus riparius (Environmental Consulting & Testing, Superior, WI) that was cultured and maintained under controlled conditions in our laboratory. The laboratory population was used for life-stage studies. The larvae used in these studies were reared from a single egg mass deposited by adult flies.
Water quality
The following water quality parameters were measured at KM, NEC, and BH: temperature (°C), pH, DO (mg l−1), salinity (ppt), and redox potential (mV). The parameters were measured using a YSI meter, model 556 (YSI Environmental, Yellow Springs, OH). Water quality parameters at RR sites include temperature (°C), pH, DO (mg l−1), and hardness (mg l−1). Hardness and DO were measured using LaMott testing kits (Carolina Science and Math, Burlington, NC). The pH was measured using a Corning pH M240 (Corning Science Products, Corning, NY).
Slide preparation of head capsules and body of chironomid
Head capsules and their corresponding body were collected for permanent mounting (Epler, Reference Epler2001) on a slide and used for species identification. The process involved decapitation of live larvae such that hemolymph was drained and collected as described below. After collection of hemolymph, the head capsule and remaining body of a particular chironomid were stored in 70% ethanol until mounting.
Species identification
All mounted head capsule samples were keyed to the lowest possible taxonomic unit based on larval body morphology and head capsule morphology keys (Epler, Reference Epler2001).
Preparation of hemolymph samples and SDS–PAGE
Hemolymph was extracted from each larva by decapitation and bleeding out onto a microscope slide. Immediately, approximately 2 μl of hemolymph was drawn up and transferred into a 1.5 ml centrifuge tube containing 14 μl of lithium dodecyl sulfide (LDS) sample buffer (Invitrogen, Carlsbad, CA), 2 μl of 8 M of urea (Qiagen, Valencia, CA), and 2 μl of 2-mercaptoethanol (Sigma Chemical Co., St Louis, MO). The samples were stored at −20 °C until use. SDS–PAGE involved separating a 5 μl aliquot of hemolymph protein mixture on 16.5% Tris–Tricine gels (BioRad, Hercules, CA) under denaturing conditions using 1X Tris/Tricine/SDS electrophoresis buffer (Biorad). All hemolymph samples in LDS sample buffer were boiled prior to loading. SeeBlue® Plus2 Protein Standard (Invitrogen) was used as a molecular weight (MW) ladder. The gel was run at 100 V followed by gel washing, fixing, staining, and drying steps. Gels were washed in ddH2O and then fixed in a fixing solution containing 50% methanol (Pharmco-Aaper, Brookfield, CT), 7% glacial acetic acid (Pharmco-Aaper), and 43% ddH20. After fixation, the gel was washed with ddH2O and stained with Gel Code Blue (Pierce, Rockford, IL). The gel was de-stained by boiled ddH2O. The gel was dried on blotting paper using a gel dryer. All gels were then scanned and imaged (see below).
Liquid chromatography–mass spectrophotometry (LC–MS) proteomic analysis
To verify that the proteins contained in particular SDS–PAGE bands were indeed Hb, four prominent bands ranging from 5 to 12.5 kDa were excised and tested using LC–MS proteomic analysis. This was done using standard protocols at the Biological Mass Spectrometry Facility of the Center for Advanced Biotechnology and Medicine of Robert Wood Johnson Medical School and Rutgers, the State University of New Jersey. The LC–MS data were searched against a subset of Uniprot database with entries containing the keyword ‘Chironomidae’ and CRAP.fasta (www.theGPM.org) and using a local version of the Global Proteome Machine (GPM cyclone, Beavis Informatics Ltd, Winnipeg, Canada).
Hb protein quantification
The gel was scanned using a Gel Doc-It Imaging Transilluminator System (UVP, Upland, CA). The picture was loaded onto VisionWorksLS program (UVP) for quantification analysis of the bands. In order to measure each band in a single lane, a 20-band system was developed using different MWs ranging from 4 to 17 kDa. Pixel intensity of each band within the 20-band system was initially quantified as a raw score and then further normalized by subtracting the background pixel intensity – a blank space in each lane. The relative intensity ratio for each band was determined by dividing the normalized pixel by the highest pixel found in that lane. Only the bands with relative intensity of 30% or greater were considered in this study as bands of interest.
Hb protein MW determination
For each hemolymph sample separated on SDS–PAGE gels, MWs were determined for all Hb bands in a single lane (Hemes, Reference Hemes1998). Relative mobility (Rf) for each protein was determined using the formula: Rf=distance of protein migration/distance of dye migration. A standard curve was generated for each gel by plotting Rf versus log scale of the MWs (log MW) of protein standards. Finally, Rf value of each Hb protein in a single lane was used to estimate its MW by interpolation to the standard curve. The ladder for SDS–PAGE contained the protein standards. Information on their MWs was provided by the manufacture's protocol (Invitrogen).
Hb protein profiles
Hb protein profiles consisted of one or more bands from a particular larva. They were associated with their corresponding head capsules, which were identified to the lowest possible taxonomic level using head capsule morphology. Hb protein profiles of all taxa were compared for uniqueness. Amino acid sequencing has shown that major Hb proteins have molecular masses in the range of 16–17.5 kDa; therefore, bands below 7 kDa were thought to be Hb protein degradation products, and were excluded from all comparisons.
Bright field imaging of chironomid head capsules
Each mounted head capsule of a chironomid was viewed using a 10X objective lens on a Zeiss Axioskop microscope (Carl Zeiss Ltd, Cambridge, UK). Images were captured using a Leica DFC 300 FX digital camera (Leica Camera Inc, Allendale, NJ). The entire head capsule could not be imaged in a single field of view using this objective, so two overlapping areas were collected (the superior and inferior portions of the head capsule) and a panoramic montage was created using the Photomerge function in Photoshop CS5 (Adobe Systems Incorporated, San Jose, CA) to generate a complete image of a head capsule. Additionally, for each area, 15–20 images from successive focal depths were collected and three-dimensional montages for each portion of the head capsule were generated using the Do Stack function in CombineZP URL (http://www.hadleyweb.pwp.blueyonder.co.uk/).
Comparison of stage-specific Hb protein profile
Chironomids from a single egg mass were collected every 3 days starting at approximately 6 days after hatching and continuing through pupation and emergence of the adult. At each time point, three Hb samples were generated. Each sample consisted of the combined hemolymph of five individuals. All individuals collected at the first time point (6 days) had visible red pigment on the thoracic segments including the head capsule, when viewed under the dissecting microscope (Parco Scientific Company, Westland, MI). Head capsules were collected from the same individuals used for hemolymph analyses. The widths of the head capsules were measured in order to determine larval instar using methods previously described (Watts & Pascoe, Reference Watts and Pascoe2000). Hemolymph from pupae and adult flies were collected the same way as from larvae. All hemolymph was separated by SDS–PAGE as described above, except that the protein concentration of all samples was first analyzed using Bicinchoninic Acid Kit (Sigma). This ensured that approximately 200 μg of total protein per sample was loaded and separated by SDS–PAGE.
Results
Water quality of the sampling sites
Water quality parameters of the four sites at the time of chironomid collection are shown in table 1. KM stood out as having water temperatures approximately 8 °C warmer than the others and supersaturated DO. The redox of KM was slightly higher as well but had a wide range, −153 to +21.9 mg l−1. These factors indicated a eutrophic environment at KM. The pH averages at KM, RR, NEC, and BH were 8.29, 7.10, 6.39, and 6.33, respectively. Salinity averaged higher at BH and KM, 2.90 and 1.71 ppt, respectively, and lower at NEC, 0.65 ppt. RR was a freshwater site and hardness averaged 180 ppm.
Chironomid head capsule identification
Sixty-six chironomids were analyzed for head capsule morphology (table 2). This included one group – Thienemannimyia and four genera – Chironomus, Cricotopus, Dicrotendipes, and Glyptotendipes. Within the four genera, there were four described species and three unidentifiable species based on head capsule morphology. Those not identified to species were denoted as ‘sp.’ or named after the region in which the chironomid was found – ‘sp. ME1’ for an unknown species collected in Maine.
Table 2. Chironomid taxonomy based on head capsule morphology. Chironomids were collected from four different sampling sites during 2007–2012. n is the total number of chironomids analyzed for a particular species. Subfamily of each species is also provided.

Site abbreviations as in Table 1.
Protein composition analysis by LC–MS
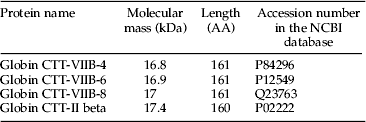
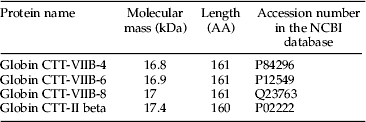
LC–MS was used to verify protein composition of the bands separated on SDS–PAGE gels. Bands at 12.5, 11, 7, and 5.5 kDa were chosen from a hemolymph sample of C. riparius (laboratory strain) for LC–MS analysis. The major proteins identified for one of the four bands (12.5 kDa) analyzed is shown in table 3. Information provided includes the protein name, molecular mass (kDa), the amino acid length, and the corresponding accession numbers in the NCBI database. According to the base peak ion chromatogram, isomers of Globin CTT Hb-VIIB (CTT-VIIB) and Globin CTT-II were the dominant proteins in the 14.5 kDa band and accounted for the majority of its intensity. CTT is the former name of C. riparius, the species used for this analysis. Similar protein species of Hbs were also observed in the 11, 7, and 5.5 kDa bands (data not shown). The LC–MS analysis indicated that the Hb proteins detected in these four bands might be breakdown products of the major Hb proteins, as the expected sizes of those Hb proteins range from 16.8 to 17.4 kDa, but no bands were found to be in this range on SDS–PAGE for C. riaprius.
Table 3. LC–MS data of the most abundantly identified proteins in Hb band 5 separated by SDS–PAGE. CTT represents Chironomus thummi thummi, which now is called Chironomus riparius. AA represents the total number of amino acids in the sequence.
Comparison of head capsule and corresponding Hb protein profile index
The head capsule of wild C. riparius and its associated Hb proteins generated by SDS–PAGE were compared (fig. 1). An important landmark of C. riparius head capsule was three dark inner teeth on the mandible (fig. 1a). According to the head capsule morphology, we determined that all individuals analyzed appeared to be the same species, regardless of the sampling sites, KM (n=2) and RR (n=4). C. riparius had three varying Hb protein profiles – P1, P2, and P3 (fig. 1b). P1 was obtained from a chironomid collected in RR, whereas P2 and P3 were from KM. All three profiles shared bands at 14.5, 12.5, and 11 kDa. Both P1 and P2 had an extra band at 11.5 kDa, and P3 showed an absence of bands at 15.5 and 11.5 kDa. P1 did not have bands below 11 kDa, whereas both P2 and P3 did.

Fig. 1. Comparison of head capsule with Hb proteins of Chironomus riparius. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profiles 1–3 (P1, P2, and P3, respectively). C. riparius has a mandible with three dark inner teeth indicated by the solid arrow (a). One species was associated with one head capsule morphology (a) and three Hb profiles (b). P1 was found in all chironomids collected in Rahway River, NJ, n=4. P2 and P3 were in chironomids collected in Kearny Marsh, NJ, n=5 for each profile.
An unidentifiable species of wild Chironomus was collected only at BH and was denoted as Chironomus sp. ME1. The head capsule of this species had two dark inner teeth on the mandible and a significantly downsized outer tooth on the mentum (fig. 2a) – traits which are atypical of C. riparius. There was only one Hb protein profile (P4) found in all individuals of Chironomus sp. ME1, n=9 (fig. 2b). P4 had three bands at 15.5, 14.5, and 11.5 kDa.

Fig. 2. Comparison of head capsule with Hb proteins of Chironomus sp. ME1. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profile 4 (P4). Distinctions between C. riparius and this unknown species are the two dark inner teeth of the mandible indicated by the solid arrow and a reduced outer tooth on the mentum indicated by the dotted arrow (a). One species associated with one head capsule morphology (a) and one Hb profile (b). This single Hb protein profile was found in all individuals of this unknown species collected at Bass Harbor, ME, n=9.
Cricotopus bicinctus was collected at RR (n=3). The most prominent feature on the head capsule was the jagged edges of the molar margin of the mandible (fig. 3a). Interestingly, C. bicinctus displayed an Hb protein profile (P5) containing only one band at 17 kDa (fig. 3b). P5 was found in all three individuals of this species.

Fig. 3. Comparison of head capsule with Hb proteins of Cricotopus bicinctus. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profile 5 (P5). The solid arrow (a) indicates the inner teeth of the mandible. One species was associated with one head capsule morphology (a) and one Hb profile (b). This single Hb protein profile was found in all individuals of this species, which was only found at Rahway River, NJ, n=3.
Dicrotendipes modestus was found at NEC (n=3), BH (n=1), and RR (n=6). The main distinguishable characteristic of this species’ head capsule was the striations on the ventromental plate: there was a mean of 32 strial ridges (fig. 4a). The Hb protein profile consisted of three distinct bands in the range of 17.5, 15, and 13.5 kDa (fig. 4b). Although D. modestus was found in three different locations, the single Hb protein profile (P6) and head capsule morphology were consistent among all individuals collected.

Fig. 4. Comparison of head capsule with Hb proteins of Dicrotendipes modestus. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profile 6 (P6). The solid arrow (a) indicates the ventromental plate. One species was associated with one head capsule morphology (a) and one Hb profile (b) in all individuals collected. D. modestus was found at three of the four sites investigated – North East Creek, ME (n=3); Bass Harbor, ME (n=1); and Rahway River, NJ (n=6).
Glyptotendipes paripes was only found at KM (n=14). Taxonomy of the genus Glyptotendipes has been under revision by Michael Heyn; however, the G. paripes has been exceptionally well characterized and is, unlike many other Glyptotendipes species, usually identifiable in all life stages. Two common features of G. paripes head capsule were the smooth anterior margin of the ventromental plate and the darkened internal area posterior to the mandible (fig. 5a). G. paripes had two similar Hb protein profiles, P7 and P8, n=11 and 3, respectively (fig. 5b). Both profiles shared two intense bands, one at 17 kDa and the other at 13 kDa. However, P8 had an extra band at 16 kDa that was not visible in P7. In addition, P8 lacked bands in the range of 11–8 kDa.

Fig. 5. Comparison of head capsule with Hb proteins of Glyptotendipes paripes. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profiles 7 and 8 (P7 and P8, respectively). Common traits of G. paripes include the smooth anterior margin of the ventromental plate indicated by the dotted arrow and the darkened internal area posterior to the mandible indicated by the solid arrow. One species was associated with one head capsule morphology (a) and two Hb protein profiles (b). Both profiles were found in chironomids collected at Kearny Marsh, NJ, n=11 for P7 and n=3 for P8.
There was an unidentifiable species of Glyptotendipes found in KM along with G. paripes (fig. 6a). Other than the 13 teeth on the mentum – a well-characterized landmark of the Glyptotendipes genus – there was a lack of characteristic traits, preventing species-level identification using head capsule morphology. The smooth anterior margin of the ventromental plate and the darkened internal area posterior to the mandible of G. paripes were not visible on this species (fig. 5a). The head capsule of Glyptotendipes sp. and its associated Hb proteins were compared (fig. 6). This unidentified species of Glyptotendipes had three varying Hb protein profiles (fig. 6b). All three profiles, P9, P10, and P11 were obtained from chironomids collected in KM, n=4, 8, and 2, respectively. All three profiles shared four bands at 17, 16, 14, and 13 kDa with varying intensity. P9 and P11 did not have bands in the range of 11 to 8 kDa, whereas P10 had multiple bands in that range. P11 had an extra band at 15 kDa that was absent in both P9 and P10.

Fig. 6. Comparison of head capsule with Hb proteins of Glyptotendipes sp. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profiles 9–11 (P9, P10, and P11, respectively). The solid arrow (a) indicates the larval mentum. One species was associated with one head capsule morphology (a) and three Hb protein profiles (b). All three profiles were found in chironomids collected at Kearny Marsh, NJ, n=4, 8, and 2, for profiles P9, P10, and P11, respectively.
The last species studied was a member of the Thienemannimyia group, a complex of several closely related genera of the subfamily Tanypodinae. Larvae of this group are difficult or impossible to identify to genus without an associated pupa or adult male. The individuals collected in this study at RR (n=2) could not be identified to the genus or species level. The head capsule of Thienemannimyia group sp. and its associated Hb proteins were compared (fig. 7). The head capsule morphology of Thienemannimyia group sp., which is typical for the subfamily Tanypodinae, was strikingly different from the other genera and species found in this investigation (fig. 7a). There was only one corresponding Hb protein profile, P12, found in the two individuals collected (fig. 7b).

Fig. 7. Comparison of head capsule with Hb proteins of Thienemannimyia group sp. Presented are (a) head capsule at 10X magnification and (b) MW ladder (L) and Hb protein profile 12 (P12). The solid arrow (a) indicates a short apical tooth on the mandible. One species was associated with one head capsule morphology (a) and one Hb protein profile (b). The Thienemannimyia group sp. individuals were collected at Rahway River, NJ, n=2.
Hb protein profile comparison
Profiles of all species were compared to determine if there was any relationship between species within a genus or among the different genera (fig. 8). Band numbers 1–20 represented all of the Hb bands found in the different profiles. The numbers matched up with MWs ranging from 17.5 to 8 kDa. Profiles of the genus Chironomus (P1–P4) shared a band at 14.5 kDa, regardless of species and collection site. Profiles of C. riparius (P1–P3) shared two more bands: 12.5 and 11 kDa. These bands might have been unique to all C. riparius individuals or only to C. riparius in NJ, where the individuals with these three profiles were collected. Cricotopus spp. (P5) had one band at 17 kDa, which was shared by Glyptotendipes spp. However, the profile of C. bicinctus (P5) only had one band at 17 kDa, whereas species of Glyptotendipes had multiple bands below 17 kDa. Thus, results indicated that a band at 17 kDa alone might identify members of the genus Cricotopus, but other Orthocladiinae must first be investigated. Dicrotendipes spp. (P6) had one consistent profile regardless of collection site. Those bands at 17.5, 15, and 13.5 kDa were not shared by other genera except the one at 15 kDa, which was shared with Glyptotendipes sp. (P11). The band at 17.5 kDa was the highest MW band observed and not found in any of the other species. The uniqueness of P6 indicated that bands in this profile could be used to identify D. modestus. Profiles of the genus Glyptotendipes (P7–P11) shared two bands at 17 and 13 kDa. Results indicated that these two bands were characteristic of the genus Glyptotendipes, since no other genera found had a band at 13 kDa. Another common band in Glyptotendipes was at 16 kDa. It was found in all profiles of the unidentified Glyptotendipes species, P9–P11, but only one of two profiles found for G. paripes. Distinguishing Glyptotendipes at the species-level may be difficult since the profiles of G. paripes (P7 and P8) were not considerably different from those of the unknown species. Thinemannimyia group sp. (P12) had one band at 14 kDa, which was also found in Glyptotendipes sp. (P9–P11).

Fig. 8. Comparison of all Hb protein profiles. Bands of each profile (P1–P12) were plotted against MW of Hb protein ranging from 17.5 to 8 kDa. A black line within a column indicates the presence of a band at that particular MW. Band intensity was not considered in this analysis. Profiles are grouped by genus.
Changes in Hb protein profile during a typical life cycle of C. riparius
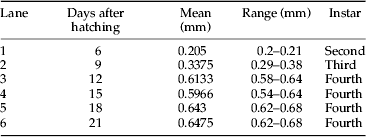
Although the Hb protein profile index corresponded well with head capsule morphology, there were multiple profiles for some species, particularly in the genera Chironomus and Glyptotendipes. These multiple profiles could complicate the use of this technique for taxonomic identification. Therefore, it was investigated whether or not developmental stage could account for this observation. To study the influence of developmental stage on Hb profiles, hemolymph was collected from a laboratory cohort of C. riparius through most of their life cycle. The hemolymph was collected at second instar (lane 1), third instar (lane 2), fourth instar (lanes 3–6), pupa (lane 8), and adult (lane 9) (fig. 9). The instar was based on average head capsule width of larvae contributing to each hemolymph sample (table 4). Head capsule widths measured in this study showed a range similar to that found by Watts & Pascoe (Reference Watts and Pascoe2000).

Fig. 9. Changes in Hb protein throughout the life cycle of a laboratory population of Chironomus riparius. Dotted box indicates 12.5 kDa band, which was found to be unique to C. riparius and observed to be continuously synthesized throughout the life cycle. Each lane presents one of three samples collected at the same time point. Lane 1: second instar, lane 2: third instar, lanes 3–6: fourth instar, lane 7: ladder, lane 8: pupa, and lane 9: adult (Table 4). Dotted black arrow indicates newly synthesized band. Black arrow indicates loss of the original 15.5 kDa band, which was present starting at fourth instar.
Table 4. Mean head capsule width (mm) for a laboratory strain of C. riparius used in a developmental study. Mean head capsule width was calculated from the five larvae whose hemolymph was combined to make a representative Hb sample (fig. 9).
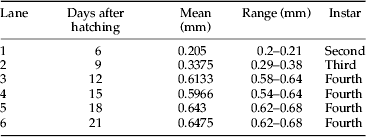
Results for the developmental study showed stage-specific changes in synthesis of Hb protein. A band at 12.5 kDa initially appeared at the late second instar (lane 1, fig. 9) and showed a perpetual synthesis throughout the larval stages that carried into both pupa and adult stages. Interestingly, a band at 12.5 kDa was also observed in P1–P3 of wild C. riparius individuals (fig. 1), indicating that this band might be unique to C. riparius species since it was not observed in the unknown Chironomus species (fig. 2). In addition, larvae at the onset of fourth instar (lane 3) showed a banding pattern similar to P3, in which a band at 15.5 kDa was missing. This finding indicated that the P3 Hb profile found in wild Chironomus might have been from an individual collected at the onset of fourth instar. Bands below 7 kDa – which were considered to be degraded Hb protein product – appeared during the third instar and remained throughout the larval stages. The distinctive profile of C. riparius was observed by the mid fourth instar (lanes 4–6), which was indistinguishable from those of P1 and P2 found in wild Chironomus (fig. 1). During the transition from larva to pupa (lanes 6–8), most of the bands were retained with the exception of bands between 4 and 6 kDa in pupa, a newly synthesized band at 16 kDa in pupa, and the disappearance of the 15.5 kDa band in adult. Synthesis of the 15.5 kDa band completely ceased in adult while the 16 kDa band became more prominent – suggesting that loss and formation of these could be due to metamorphosis.
Discussion
Species identification becomes crucial when changes in communities are used to study biodiversity and to biomonitor species-level responses to anthropogenic stressors. Chironomids are important members of the BMI community and an easier method for their identification would facilitate their use in field studies. The approach of this study was to compare a new technique of identification, SDS–PAGE of Hb proteins, with a well-established one, larval head capsule morphology. Identification of chironomids using SDS–PAGE was validated by showing that a unique combination of Hb protein sizes was associated with just one head capsule morphology and by proving LC–MS that the bands from SDS–PAGE gels were actually composed of Hb proteins.
In this study, Hb protein profiles enhanced the ability to identify chironomid species. For example, individuals of C. bicinctus, Thienemannimyia group sp., and D. modestus showed distinctive Hb profiles with no variation. However, C. bicinctus and Thienemannimyia group sp. were only found at one site, RR, and only two to three individuals of each species were analyzed. Studying more individuals from more locations may increase the number of Hb profiles for these two species. On the other hand, D. modestus found at three different locations had three distinct bands – making a consistent, unique profile that was not found in any other species.
Two genera, Chironomus and Glyptotendipes, had more than one Hb profile associated with each head capsule. This complicated the use of their profiles for species identification. C. riparius and both species of Glyptotendipes were found in KM. Due to shared bands, a unique profile for each genus was not found, although they did have characteristic bands such as 12.5 kDa in C. riparius (P1–P3) and 17 and 13 kDa in both Glyptotendipes species (P7–P11). Since chironomids have been shown to secrete stage-specific Hbs (Vafopoulou-Mandalos & Laufer, Reference Vafopoulou-Mandalos and Laufer1982, Reference Vafopoulou-Mandalos and Laufer1984), a developmental study was undertaken to determine if one or more of the Hb bands in the different profiles could be used to consistently identify the species (fig. 9). The study was performed using a laboratory population of C. riparius. Results showed one consistent band at 12.5 kDa that was first observed at second instar and continued to be observed throughout the whole life cycle. This indicated that despite differences in stage-specific profiles the presence of this band could be used to identify C. riparius. This was supported by the absence of this band in the unknown Chironomus species (fig. 2) as well as the rest of the other wild chironomids collected (fig. 8).
Results from the developmental study appeared to account for the variable profiles seen in wild C. riparius (P1–3). The Hb protein profile for P3 was missing the 15.5 kDa band which was found in both P1 and P2, indicating that the P3 individual might have been transitioning between the third and fourth instar (compare lanes 3 and 4 in fig. 9). This finding suggested that while head capsule width could be used to identify larval instars, Hb protein profile might be another tool once it is fully characterized for a particular species. Findings also indicated that it would be beneficial to analyze multiple instars of a particular species to determine which band or combination of bands are consistently present and therefore representative of the species.
Many studies have shown that environmental stress can influence chironomids at both the molecular and cellular levels (Lee et al., Reference Lee, Lee, Park and Choi2006; Ha & Choi, Reference Ha and Choi2008; Nair et al., Reference Nair, Park and Choi2011). Findings in this study suggested that water chemistry might have contributed to Hb polymorphism detected by multiple Hb protein profiles for some species. For example, two major differences between C. riparius at RR and KM were the absence of the 8.5 kDa band in RR chironomids as well as the presence of bands below 7 kDa in KM chironomids. These differences might be attributed to salinity given that RR is freshwater while KM is oligohaline. This idea was supported by the presence of bands below 7 kDa in C. ME1, which was collected at another oligohaline site. The modifications in Hb protein seen at oligohaline sites might reflect a role in salt tolerance. Research has suggested that C. salinarius uses their hemolymph for osmotic regulation (Cartier et al., Reference Cartier, Claret, Garnier and Franquet2011). C. salinarius appeared to absorb and eliminate excess salt using their Hb proteins or were able to adjust their intracellular Hb protein levels according to the external environment. Interestingly, the three polymorphic species found in this study – Chironomus, Glyptotendipes, and Dicrotendipes – were all found at oligohaline sites. In addition, the two taxa that showed a lack of polymorphism, C. bicinctus and Thienemannimyia group sp., were found only at freshwater sites. This limited evidence suggested that polymorphism in Hb proteins may be an important feature of chironomid adaptation to environmental parameters.
Species-specific digestion of Hb proteins might have accounted for the different band sizes found in chironomid. Results of LC–MS analysis showed that proteins found in the band with MW of 12.5 kDa were partial products or fragments of mainly Hb genes VII and II (table 3). Since the full-length protein products of these Hb genes have a MW of approximately 17 kDa, the Hb proteins in the band at 12.5 kDa and others (with 15.5 kDa being the highest) must have been altered so as to reduce their MWs. Research has shown that proteins are degraded by a number of natural processes (Mann & Jensen, Reference Mann and Jensen2003; Jensen, Reference Jensen2004). Theoretically, the stage of larval development and or responses to ecological conditions could have affected the activation of specific protease(s) such as ubiquitin and resulted in a variety of low MW proteins or even the absence of proteins. Support for this hypothesis was that the only difference in some Hb profiles was the presence or absence of lower bands. This can be seen by comparing P7 and P8 of G. paripes and P1 and P2 of C. riparius. Other reasons for changes in Hb proteins have included seasonal changes (Ribbing & Ruterjans, Reference Ribbing and Ruterjans1980; Osmulski & Leyko, Reference Osmulski and Leyko1986) and gradual increases in Hb protein concentration from early to later instars (Schin et al., Reference Schin, Poluhowich, Gamo and Laufer1974; Bergtrom et al., Reference Bergtrom, Laufer and Rogers1976). Taken together, these studies support the idea that there are natural processes for modifying Hb protein concentrations and profiles in hemolymph. This variation in Hb digestion products could contribute to taxonomic identification as Hb is physiologically relevant and likely under selective environmental pressure.
This study contributes to the development of a novel means of identification of wild chironomid species using Hb protein profiles detected by SDS–PAGE. The technique described here supports and supplements the current standard method of taxonomic identification using larval head capsule morphology or even other methods such as PCR-based approaches described above. Identification using Hb profiles requires less taxonomic expertise, could be used to identify larvae with deformed head capsules, and most importantly, could handle large numbers of samples. The consistent presence of particular bands for each species studied indicates that Hb profiles could be used to study taxonomic relationships and determine instars in chironomids. Since more than one profile was found for some species, the effect of environmental conditions and or stressors might also be studied using this technique.
Acknowledgements
We sincerely thank Dr Angela Klaus at Seton Hall University for her guidance and technical support with use of microscopes and image processing. We are grateful to Dr Haiyan Zheng at the Biological Mass Spectrometry Facility of the Center for Advanced Biotechnology and Medicine of Robert Wood Johnson Medical School and Rutgers, the State University of New Jersey for her valuable technical support with LC–MS. The anonymous reviewers are also appreciated for their time and effort. Dr Carolyn S. Bentivegna was supported by a Mount Desert Island Biological Laboratory, New Investigator Award, 2008 – when collecting chironomids from Maine and by an award from the Meadowlands Environmental Research Institute, 2004–2009, when collecting chironomids from Kearny Marsh, NJ.
Author's contributions
C.S.B. conceived the research collected most of the material. C.S.B and J.T.O mounted the head capsules of chironomids. The representative specimens were identified by J.H.E. J.T.O conducted the laboratory work including microscopic imaging development and analysis of the data and J.T.O wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.