Introduction
Malaria is a parasitic disease that has historically threatened human health, particularly in tropical and sub-tropical regions (Zhou et al., Reference Zhou, Zhang, Yang, Liu, Zhao, Li, Qian and Xu2016). It is considered as one of the three major global public health problems, together with HIV/AIDS and tuberculosis (Vitoria et al., Reference Vitoria, Granich, Gilks, Gunneberq, Hosseini, Were, Raviqlione and De Cock2009). In early 2016, malaria was reported to be endemic in 91 countries and territories, and about 212 million cases and 429 000 deaths occurred globally in 2015 (WHO, 2016), of which more than 90% were in Africa. In 2010, the government of China announced its plan to eradicate malaria in the entire country by the end of 2020. The action plan was thus launched that same year in Henan Province. With the progress of malaria elimination, no local malaria cases have been reported in Henan Province since 2012 (Liu et al., Reference Liu, Zhang, Zhou, Yang, Qian, Zhao and Xu2014a). However, with increasing travel, migration, economic globalization and international exchanges, malaria, especially Plasmodium falciparum, has re-emerged as a major public health challenge (Feng et al., Reference Feng, Yan, Feng, Zhang, Li, Xia and Xiao2014; Liu et al., Reference Liu, Hsiang, Zhou, Wang, Cao, Gosling, Cao and Gao2014b), with over 90% of cases originating from Africa (Liu et al., Reference Liu, Zhou, Qian, Yang and Zhang2014c; Zhang et al., Reference Zhang, Zhou, Feng, Fang and Xia2015; Yang et al., Reference Yang, Qian, Chen, Liu, Zhou and Zhang2016).
A total of 212 million malaria cases were reported in 2015, reflecting a 14% decline since 2010 and 22% since 2000. The observed decrease in malaria prevalence was mainly attributable to the application of efficacious antimalarial drugs. Among these, chloroquine (CQ) has been proven to be one of the most effective drugs against malaria, particularly in the highly endemic regions of Africa (Wellems and Plowe, Reference Wellems and Plowe2001). The success of CQ and its extensive use have led to the development of resistance in P. falciparum from the late 1950s (Maberti, Reference Maberti1960; Moore and Lanier, Reference Moore and Lanier1961; Reyes, Reference Reyes1981; Peters, Reference Peters1987). CQ resistance was first reported in 1957 at the Thailand–Cambodia border, which eventually spread to other countries around the world (Wernsdorfer and Payne, Reference Wernsdorfer and Payne1991; Mehlotra et al., Reference Mehlotra, Fujioka and Roepe2001; Ridley, Reference Ridley2002). Artemisinin combination therapies (ACTs) were then recommended against uncomplicated P. falciparum infections in 2006 (WHO, 2006). ACTs were considered to be the most effective treatment for P. falciparum. However, in 2008, artemisinin-resistance strains were reported in Western Cambodia, followed by cases in Southeast Asia and the Great Mekong Subregion (GMS) (Muller et al., Reference Muller, Sie, Meissner, Schirmer and Kouyate2009; Wang et al., Reference Wang, Shrestha, Li, Miao, Yuan, Cabrera, Grube, Yang and Cui2015).
Pfmdr1, a gene located on P. falciparum chromosome 5, encodes P-glycoprotein homologue-1, which is a molecular marker for a variety of antimalarial drugs. Mutations involving codons 86, 184, 1034, 1042 and 1246 have been shown to be strongly linked to a variety of antimalarial drug resistances, including artemisinin (Guan et al., Reference Guan, Tang, Hu, Feng and Liu2005; Gama et al., Reference Gama, Pereira-Carvalho, Lutucuta Kosi, Almeida de Oliveira, Fortes, Rosenthal, Daniel-Ribeiro and Ferreira-da-Cruz2010). In Africa, resistance to CQ, amodiaquine (AQ) and antifolate antimalarial drugs has been observed (Trape et al., Reference Trape, Pison, Spiegel, Enel and Rogier2002; Roper et al., Reference Roper, Pearce, Bredenkamp, Gumede, Drakeley, Mosha, Chandramohan and Sharp2003), and a decreased response to artesunate (AS) plus AQ has also been reported (Nsobya et al., Reference Nsobya, Dokomajilar, Joloba, Dorsey and Rosenthal2007). Although there was no evidence of artemisinin resistance in Africa, its impending spread was predicted to be disastrous (Mok et al., Reference Mok, Imwong, Mackinnon, Sim, Ramadoss, Yi, Mayxay, Chotivanich, Liong, Russell, Socheat, Newton, Day, White, Preiser, Nosten, Dondorp and Bozdech2011). In Henan Province, all registered malaria cases since 2012 were imported, with over 90% involving migrant workers from Africa. It is thus essential to understand the distribution of antimalarial drug resistance to assist in the development of prevention, control and elimination schemes for malaria in Henan Province. Therefore, this study was designed to evaluate drug resistance marker polymorphisms in migrant workers in Henan Province to provide rational suggestions to prevent and treat future cases. In this study, the prevalence of polymorphisms in the Pfmdr1 gene was determined from P. falciparum-positive African patients in Henan Province from 2012 to 2016.
Materials and methods
Sample collection and DNA extraction
Blood samples were collected from P. falciparum-infected African migrant workers prior to treatment and were labelled with study numbers, names and dates. All patients returned from Africa to Henan Province in 2012–2016. The patients were finally diagnosed by microscopic examination of Giemsa-stained thick and thin blood smears and nested polymerase chain reaction (PCR). All the blood samples were stored at −20 °C until analysis. DNA was isolated from the blood samples using the QIAamp DNA Mini kit (QIAGEN Inc., Frankfurt, Germany) according to the manufacturer's instructions and stored at −20 °C until PCR analysis.
Pfmdr1 gene amplification and sequencing
Amino acid substitutions N86Y, Y184F, S1034C, N1042D and D1246Y in the Pfmdr1 gene were screened. Polymorphisms were evaluated using nested PCR. The primary round of PCRs was performed in a total volume of 25 µL, which consisted of the following: 8.5 µL of ddH2O, 1 µL each of first-round primer (10 µmol L−1), 12.5 µL of a 2 × Go Taq Green Master Mix (PROMEGA Inc., Madison, WI, USA) and 2 µL of the DNA template. The PCR reaction conditions were as follows: 95 °C for 3 min, followed by 30 cycles of 95 °C for 30 s, 54 °C for 45 s and 72 °C for 30 s; and a final extension of 72 °C for 5 min. The second round contained the second-round primers and 1.5 µL of the first-round PCR product as template, with the same cycling conditions as that described for the first round. The primer sequences were shown in Table 1. Sequencing was conducted by Shanghai DNA BioTechnologies Co., Ltd. (Shanghai, China).
Table 1. PCR primer sequences used for the amplification sequence encoding pfmdr1

Sequencing alignments and data analysis
Sequences were analysed using BLAST (http://blast.ncbi.nlm.nih.gov/) and aligned to reference sequence PF3D7_0523000 using BioEdit Sequence Alignment Editor. The data were analysed using Microsoft Excel and SPSS 17.0. Pfmdr1 allele frequencies were calculated with Microsoft Excel and differences were assessed using SPSS 17.0. Pearson's χ 2 test was used to determine the significance of the results. P values were calculated and considered to be statistically significant at P < 0.05.
Results
Epidemiological characteristics of patients
A total of 632 isolates were collected from returning P. falciparum-infected African migrant workers in Henan Province in 2012–2016. The male:female ratio was 125.4:1 (627/5). The mean age was 38.09 ± 9.34 years (range: 17–70), of which only five patients were older than 60 years and one was <18 years of age. The 632 patients returned from 29 countries in Africa, with the majority (37.18%, 235/632) returning from West Africa, followed by South Africa (28.64%, 181/632) and Central Africa (26.58%, 168/632). Only a minority of the patients came back from East Africa and North Africa, accounting for 5.86% (37/632) and 1.74% (11/632) of the total number of patients, respectively (Table 2).
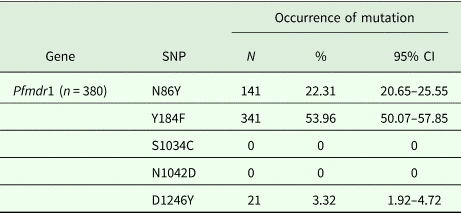
Table 2. The polymorphisms of pfmdr1 gene

SNP, single-nucleotide polymorphism; CI, confidence interval.
Screening for Pfmdr1 mutations and genotyping
The amplicon of locus 86 and 184 was 526 bp in length, and the amplicon including locus 1034, 1042 and 1246 was 799 bp in length. The amplicons were successfully sequenced from all samples. Positions 1034 and 1042 of the Pfmdr1 gene were all wild-type. Mutations involving codons 86, 184 and 1246 of the Pfmdr1 gene were identified in 380 patients. The mutation Y184F was the predominant change (53.96%) (Table 2), and 13 haplotypes were identified, including wild-type haplotype NYSND (39.87%, 252/632), three single-mutant haplotypes YYSND (2.85%, 18/632), NFSND (31.01%, 196/632) and NYSNY (0.47%, 3/632), three double-mutant haplotypes YFSND (13.77%, 87/632), NFSNY (0.32%, 2/632) and YYSNY (2.06%, 13/632), one triple-mutant haplotype YFSNY (0.16%, 1/632) and five mixed haplotypes N/Y YSND (1.90%, 12/632), N Y/F SND (6.17%, 39/632), N/Y Y/F SND (0.47%, 3/632), YYSN D/Y (0.16%, 1/632) and N/Y FSND (0.79%, 5/632). NFSND was the predominant haplotype (51.58%, 196/380, Table 3 and Fig. 1). The sequences of the mutations were deposited in GenBank under accession number MH266472-MH266479.

Fig. 1. Codons 86, 184, 1034, 1042 and 1246 haplotypes in the pfmdr1 gene of P. falciparum samples from Africa migrant workers in Henan Province. (A) The wild-type haplotype. The haplotype NYSND of sample 3D7. (B)–(H) The mutant haplotypes. The haplotypes were named YYSND, NFSND, NYSNY, YFSND, NFSNY, YYSNY and YFSNY, respectively. (I)–(M) The mixed haplotypes. The haplotypes were named N/Y YSND, N Y/F SND, N/Y Y/F SND, YYSN D/Y and N/Y FSND. Mutation are indicated by arrows.
Table 3. Distribution of Pfmdr1 polymorphisms from Africa-imported cases

Distribution of the Pfmdr1 haplotypes
Thirteen haplotypes encompassing codons 86, 184, 1034, 1042 and 1246 of the Pfmdr1 gene were identified in this study. Only one patient who returned from Sierra Leone in West Africa harboured the YFSNY haplotype. The wild-type haplotype NYSND was detected in patients originating from various regions of Africa. The other haplotypes were found in patients from two, three or four regions of Africa. Eleven isolates from patients who returned from two countries in North Africa carried three haplotypes: NYSND (72.73%, 8/11), YFSND (9.09%, 1/11) and YYSNY (27.27%, 3/11). Approximately 181 samples from patients originating from four countries in South Africa carried the wild-type haplotype NYSND (61.33%, 111/181), three single-mutant haplotypes YYSND (8.29%, 15/181), NFSND (22.65%, 41/181) and NYSNY (0.55%, 1/181), two double-mutant haplotypes YFSND (1.66%, 3/181) and NFSNY (0.55%, 1/181), and three mixed haplotypes N/Y YSND (0.55%, 1/181), N Y/F SND (3.87%, 7/181) and N/Y FSND (0.55%, 1/181). Around 37 patients from five countries in East Africa carried eight haplotypes, including the wild-type haplotype NYSND (27.03%, 10/37), two single-mutant haplotypes NFSND (48.65%, 18/37) and NYSNY (2.70%, 1/37), two double-mutant haplotypes YFSND (2.70%, 1/37) and NFSNY (2.70%, 1/37), and three mixed haplotypes N/Y YSND (2.70%, 1/37), N Y/F SND (10.81%, 4/37) and N/ Y FSND (2.70%, 1/37). Approximately 235 isolates from patients originating from 11 countries in West Africa carried 10 haplotypes, including the wild-type haplotype NYSND (28.94%, 68/235), two single-mutant haplotypes YYSND (0.43%, 1/235) and NFSND (39.14%, 92/235), two double-mutant haplotypes YFSND (18.72%, 44/235) and YYSNY (2.13%, 5/235), one triple-mutant haplotype YFSNY (0.43%, 1/235), and four mixed haplotypes N/Y YSND (2.13%, 5/235), N Y/F SND (7.23%, 17/235), YYSN D/Y (0.43%, 1/235) and N/Y FSND (0.43%, 1/235). Approximately 168 isolates from patients from seven countries in Central Africa carried 11 haplotypes, including the wild-type haplotype NYSND (33.33%, 56/168), three single-mutant haplotypes YYSND (1.19%, 2/168), NFSND (26.79%, 45/168) and NYSNY (0.59% 1/168), two double-mutant haplotypes YFSND (22.62%, 38/168) and YYSNY (2.98%, 5/168), and five mixed haplotypes N/Y YSND (2.98%, 5/168), N Y/F SND (6.55%, 11/168), N/Y Y/F SND (1.79%, 3/168), YYSN D/Y (0.59%, 1/168) and N/Y FSND (1.19%, 2/168). The highest frequency of the wild-type haplotype NYSND was observed in patients from North Africa (63.64%, 7/11), followed by South Africa (61.33%, 111/181), Central Africa (33.33%, 56/168), West Africa (28.94%, 68/235) and East Africa (27.03%, 10/37). Significant differences among groups were observed (χ 2 = 54.605, P < 0.05). NFSND was the predominant mutant haplotype in patients from East Africa, accounting for 48.65% (18/37) of the total number of haplotypes, followed by those from West Africa (39.14%, 92/235), Central Africa (26.79%, 45/168), South Africa (22.65%, 41/181) and North Africa (9.09%, 1/11). Significant differences among groups were observed (χ 2 = 22.368, P < 0.05).
Distribution of Pfmdr1 mutations based on year
From 2012 to 2016, the frequency of mutations involving codon 86 was 34.44% (31/90), 29.46% (38/129), 22.54% (32/142), 20.61% (27/131) and 9.29% (13/140), respectively, and differences among frequencies were statistically significant (χ 2 = 25.372, P < 0.05). From 2012 to 2016, the frequency of mutations involving codon 184 was 64.44% (58/90), 54.26% (70/129), 51.41% (73/142), 60.31% (79/131) and 43.57% (61/140), respectively, with significant differences among the years (χ 2 = 12.564, P < 0.05). The frequency of mutations involving codon 1246 was 2.22% (2/90), 3.88% (5/129), 4.93% (7/142), 2.29% (3/131) and 2.86% (4/140), respectively, with no significant differences among years (χ 2 = 2.133, P > 0.05) (Table 4).
Table 4. The mutations of Pfmdr1 reported in 2012–2016

Discussion
Drug resistance in P. falciparum is a global problem that has severely affected malaria control. To design an effective scheme for malaria control, it is essential to understand the patterns and distribution of antimalarial drug resistance. Although molecular markers do not directly reflect drug resistance in the malaria parasite, these may be utilized in early surveillance of alleles that undergo selection pressure. Molecular markers are currently the most convenient and efficient methods in resistance monitoring.
The volume of trade and services between China and other countries has increased in recent years. Malaria cases in Henan Province involving migrant workers have thus markedly increased, although no local cases of malaria have been reported in Henan Province since 2012. Many of the malaria cases are imported cases of P. falciparum, most of which are from Africa. The management of imported cases has become the main task for malaria control in Henan Province. The use of molecular markers for drug-resistance monitoring will provide data that may be used for the management of imported malaria cases.
The Pfmdr1 gene has been proposed as a molecular marker for CQ resistance, and it has also been reported to be associated with in vitro responses to AS and AQ (Nsobya et al., Reference Nsobya, Dokomajilar, Joloba, Dorsey and Rosenthal2007; Danquah et al., Reference Danquah, Coulibaly, Meissner, Petruschke, Muller and Mocken-haupt2010; Lin et al., Reference Lin, Juliano and Wongsrichanalai2010). In our study, Pfmdr1 polymorphisms were identified in imported P. falciparum isolates collected during 2012–2016 from migrant workers returning to Henan Province from Africa. The majority of the isolates (77.69, 100, 100, and 96.68%) carried wild-type N86, S1034, N1042 and D1246 at codons 86, 1034, 1042 and 1246, respectively, which agreed with that previously reported in isolates from the China–Myanmar border (Huang et al., Reference Huang, Tang, Yang, Zhou, Liu, Li and Guo2012) and Africa (Thomas et al., Reference Thomas, Ndir, Dieng, Mboup, Wypij, Maguire and Wirth2002; Feng et al., Reference Feng, Li, Yan, Feng and Xia2015). The high frequency of the Pfmdr1 N86Y allele in Africa has been attributed to the extensive use of AS plus AQ (Dokomajilar et al., Reference Dokomajilar, Lankoande, Dorsey, Zongo, Ouedraogo and Rosenthal2006; Froberg et al., Reference Froberg, Jornhagen, Morris, Shakely, Msellem, Gil, Bjorkman and Martensson2012). The percentage of samples carrying the mutation 184F at codon 184 was 53.96%, which was similar to the findings in Osogbo, Nigeria (Ojurongbe et al., Reference Ojurongbe, Ogungbamigbe, Fagbenro-Beyioku, Fendel, Kremsner and Kun2007).
The distribution of Pfmdr1 polymorphisms is related to geographic locations (Xu et al., Reference Xu, Wei, Li, Xiao, Yin, Kong, Wang, Cui, Sun, Zhao, Zhu, Yan and Huang2016). N86Y has been mainly reported in Asia and Africa, whereas N1042D and D1246Y occur more frequently in South America (Thomas et al., Reference Thomas, Ndir, Dieng, Mboup, Wypij, Maguire and Wirth2002; Lucchi et al., Reference Lucchi, Komino, Okoth, Goldman, Onyona, Wiegand, Juma, Shi, Barnwell, Udhayakumar and Kariuki2015). The results of this study coincided with the lower frequency of N1042D and D1246Y mutations in Africa. Thirteen genotypes were detected in our study. The wild-type NYSND was the predominant haplotype (39.87%), followed by NFSND (31.01%) and YFSND (13.77%), whereas the other haplotypes showed low frequencies. There was the largest proportion of the mutant type in West Africa, accounting for 60.85% (143/235) (YYSND, 1/235, 0.43%; NFSND, 92, 39.14%; YFSND, 44, 18.72%; YYSNY, 5/235, 2.13%; and YFSNY, 1/235, 0.43%), this difference might be due to the amount of AQ used in different regions. In this study, 9.49% of the mixed-type (wild-type and mutant) Pfmdr1 was found in falciparum malaria isolates from patients who returned from 17 countries (Zambia, Angola, Kenya, Uganda, Tanzania, Mali, Liberia, Ghana, Sierra Leone, Nigeria, Guinea, Central African Republic, Gabon, Congo, DRC, Congo, Cameroon and Equatorial Guinea). This result confirmed the widespread distribution of Pfmdr1 in Africa. The mixed haplotype of Pfmdr1 has been reported in Angola, Sudan, Nigeria, Ghana and Mozambique (Plucinski et al., Reference Plucinski, Talundzic, Morton, Dimbu, Macaia, Fortes, Goldman, Lucchi, Stennies, MacArthur and Udhayakumar2015; Xu et al., Reference Xu, Wei, Li, Xiao, Yin, Kong, Wang, Cui, Sun, Zhao, Zhu, Yan and Huang2016). This study enriched the distribution of the mixed type of Pfmdr1. Among 632 isolates, only one patient, who returned from Sierra Leone, harboured the YFSNY haplotype.
The mutant type of Pfmdr1 was shown to be associated with resistance to lumefantrine in vitro and in vivo studies (Sisowath et al., Reference Sisowath, Ferreira, Bustamante, Dahlstrom, Martensson, Bjorkman, Krishna and Gil2007; Mwai et al., Reference Mwai, Kiara, Abdirahman, Pole, Rippert, Diriye, Bull, Marsh, Borrmann and Nzila2009; Conrad et al., Reference Conrad, LeClair, Arinaitwe, Wanzira, Kakuru, Bigira, Muhindo, Kamya, Tappero and Greenhouse2014; Henriques et al., Reference Henriques, Hallett, Beshir, Gadalla, Johnson, Burrow, Schalkwyk, Sawa, Omar, Clark, Bousema and Sutherland2014). Mutations in Pfmdr1 were also considered to be associated with an increase in resistance to AS; therefore, it is valuable to monitor the resistance in various regions (Van Tyne et al., Reference Van Tyne, Dieye, Valim, Daniels, Sene, Lukens, Ndiaye, Bei, Ndiaye, Hamilton, Ndir, Mboup, Volkman, Wirth and Ndiaye2013). Henan Province implemented an action plan for malaria elimination in 2010, and no local malaria cases have been reported since 2012; over 90% of the current cases involve migrant workers returning from Africa. Angola, Nigeria and Equatorial Guinea became the top three countries from which the patients returned (Yang et al., Reference Yang, Qian, Chen, Liu, Zhou and Zhang2016; Zhou et al., Reference Zhou, Zhang, Yang, Liu, Zhao, Li, Qian and Xu2016). In this study, 632 patients returned from 29 countries in Africa, and half of them returned from Angola (145), Nigeria (87) and Equatorial Guinea (81). Surveillance and population studies are essential for the early detection and subsequent prevention of the spread of drug resistance (Mvumbi et al., Reference Mvumbi, Kayembe, Situakibanza, Bobanga, Nsibu, Mvumbi, Melin, Mol and Hayette2015), and it is very important for the goal of eliminating malaria in Henan Province.
There was a trend of increasing prevalence of isolates with wild-type genotypes at these codons, including 86 and 184, from the year 2012 to 2016. It suggested that the resistance of P. falciparum to CQ might be reduced, and sensitivity might again be achieved. In this study, the prevalence of the combination of both mutations (86Y + 1246Y) was 2.06% in the parasite population. Based on this relatively low prevalence, currently, the use of ACTs as a first-line treatment is effective to treat P. falciparum in Henan Province. These data should help provide a more comprehensive picture of antimalarial resistance and guide decisions regarding treatment policies in Henan Province.
Conclusions
This study reported the prevalence of polymorphisms in the Pfmdr1 gene in returning African migrant workers in Henan Province. The frequencies of Pfmdr1 N86Y and Y184F alleles in the samples from Africa were relatively high, whereas the mutant frequencies of S1034C, N1042D and D1246Y were low. The frequency of isolates with mutations at codons 86 and 184 decreased from 2012 to 2016. It was essential to assess the evolution of antimalarial drug resistance in returning migrant workers in Henan Province, and these data will be useful in the development of an effective treatment policy in Henan Province.
Acknowledgements
The authors thank the workers from City level CDC, who participated in the study.
Financial support
This work was supported by the Special Funding of the Henan Health Science and Technology Innovation Talent Project (No.4045). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of interest
None.
Ethical standards
The study was reviewed and approved by the Special Funding of the Henan Health Science and Technology Innovation Talent Project (No. 4045). Before sampling, the purpose of the research study was presented to the participants, and consent was obtained from those who agreed to participate in the investigation.