Introduction
High-yielding varieties (HYVs) and elite germplasm derived from semi-dwarf rice breeding since the 1960s have been applied either on a large scale in rice production or as parents in developing new inbred and hybrid rice varieties. Germplasm exchange among breeding programmes promotes new variety development, but it may also increase genetic similarity as all breeding programmes pursue limited common variety traits.
Crop heterosis is significantly increased by combining genetically distinct parental materials, i.e., parents belonging to distinct heterotic pools. Heterotic pools can be identified through the study of agro-morphological differentiation and the characterization of genetic diversity at the DNA level via molecular markers. Detection of DNA polymorphisms has been extensively employed as a mean to assess genetic diversity and its relationship to heterosis in maize (Lee et al., Reference Lee, Godshalk, Lamkey and Woodman1989; Dudley et al., Reference Dudley, Saghai Maroof and Rufener1991; Boppenmaier et al., Reference Boppenmaier, Melchinger, Seitz, Geiger and Herrmann1993; Reif et al., Reference Reif, Melchinger, Xia, Warburton, Hoisington, Vasal, Srinivasan, Bohn and Frisch2003) and rice (Xie, Reference Xie1993; Zhang et al., Reference Zhang, Gao, Yang, Ragab, Saghai Maroof and Li1994, Reference Zhang, Gao, Saghai Maroof, Yang and Li1995). Early studies have shown that heterotic rice hybrids in China were derived from distant parents of different geographical origins or ecotypes or subspecies (Yuan, Reference Yuan1977). Two heterotic pools, i.e., early-season indica varieties in the regions of the Yangtze River Valley and mid- or late-season indica varieties from South and Southeast Asia (mainly the varieties developed at the International Rice Research Institute (IRRI) in the Philippines) were identified for the three-line Chinese hybrid rice (Yuan, Reference Yuan1977; Lu and Xu, Reference Lu and Xu2010) with many commercial hybrids developed. Parental selection based on distant relations geographically and biotype differences have been the most important factors for heterotic hybrid development, but the recent development of super high-yielding hybrids in China has involved many crosses between subspecies or parents derived from subspecies. The development of heterotic rice hybrids still relies mostly on the breeders' trial-and-error approach. It is both urgent and justified to explore the genetic relationships and diversity of parental populations more widely and in more detail for further hybrid rice breeding. The study of genetic variation among southern Chinese and IRRI rice varieties attracts the interest of rice breeders since indica is the major cultivated rice in southern China and in other tropical Asian countries, and it provides a rich source of germplasm as hybrid rice parents.
Detection and analysis of genetic diversity in rice germplasm have been immensely facilitated with the use of molecular markers (Collard et al., Reference Collard, Vera Cruz, McNally, Virk and Mackill2008; Moose and Mumm, Reference Moose and Mumm2008). Single nucleotide polymorphisms (SNPs) are widely becoming the marker of choice due to their high-throughput capacity, cost-effectiveness, efficiency, and abundant type of polymorphisms to potentially increase the speed and efficiency of research and breeding activities (McNally et al., Reference McNally, Childs, Bohnert, Davidson, Zhao, Ulat, Zeller, Clark, Hoen, Bureau, Stokowski, Ballinger, Frazer, Cox, Padhukasahasram, Bustamante, Weigel, Mackill, Bruskiewich, Ratsch, Buell, Leung and Leach2009; McCouch et al., Reference McCouch, Zhao, Wright, Tung, Ebana, Thomson, Reynolds, Wang, DeClerck, Ali, McClung, Eizenga and Bustamante2010; Chen et al., Reference Chen, He, Zou, Chen, Yu, Liu, Yang, Gao, Xu, Fan, Li, Li and Deng2011; Thomson et al., Reference Thomson, Zhao, Wright, McNally, Rey, Tung, Reynolds, Scheffler, Eizenga, McClung, Kim, Ismail, Ocampo, Mojica, Reveche, Dilla-Ermita, Mauleon, Leung, Bustamante and McCouch2012). SNP marker profiling of rice germplasm has shown great genetic variability and significant differentiation among rice subspecies or landraces (Zhao et al., Reference Zhao, Tung, Eizenga, Wright, Ali, Price, Norton, Islam, Reynolds, Mezey, McClung, Bustamante and McCouch2011), yet none of the documented studies has so far dealt with genetic diversity in HYVs and their potential application for heterotic pools.
In this study, using SNP markers, we evaluated the population structure of southern Chinese and IRRI rice varieties that have been popularly cultivated on a large scale of production and/or are also used extensively in rice breeding programmes as donor parents, but, most importantly, those varieties represent the two known rice heterotic pools. This study aims to characterize and analyse the population structure and diversity based on SNP molecular markers, and to provide baseline data and references for parental selection to further the studies on rice heterotic pools.
Materials and methods
Plant materials
Plant materials for the study comprised 215 diverse Oryza sativa L. indica varieties selected from 11 provinces in southern China (161) and IRRI (54). The Chinese samples were from the rice germplasm bank at the China National Rice Research Institute (CNRRI) and originated from Anhui (6 varieties), Fujian (17), Guangdong (33), Guangxi (6), Henan (5), Hubei (6), Hunan (18), Jiangsu (3), Jiangxi (23), Sichuan (17) and Zhejiang (27). The IRRI samples were the lowland irrigated inbreds historically released as commercial varieties, including from IR8 (released in 1966) to IRRI154 (released in 2009) (see Table S1, available online only at http://journals.cambridge.org). All of the CNRRI and IRRI varieties were both commercial rice cultivars grown on a large scale and parents that were widely used in various rice breeding programmes in China and in South and Southeast Asian countries. Information on all the IRRI varieties is available from the IRRI database (http://www.iris.irri.org/germplasm/) while the information on all the Chinese varieties can be requested from the website http://chinariceinfo.com or http://icgr.caas.net.cn.
DNA extraction and SNP genotyping
All of the samples were cleaned and purified using single-seed-descent in the previous generations. Genomic DNA from the samples was isolated from three to five leaves of 21-d-old plants per line using the modified cetyltrimethylammonium bromide method (Murray and Thompson, Reference Murray and Thompson1980). The isolated DNA was RNase-treated and the final concentration was normalized to 50 ng/μl. The GoldenGate Genotyping Protocol of the Veracode Technology was applied in SNP genotyping, which was carried out at the Molecular Marker Applications Laboratory at IRRI. RiceOPA2.1, a 384-plex oligonucleotide pool assay (OPA) designed for indica/indica genotypes (Thomson et al., Reference Thomson, Zhao, Wright, McNally, Rey, Tung, Reynolds, Scheffler, Eizenga, McClung, Kim, Ismail, Ocampo, Mojica, Reveche, Dilla-Ermita, Mauleon, Leung, Bustamante and McCouch2012), was used as markers.
SNP data generation and analysis
Scan results were generated from Illumina's BeadXpress Reader, from which the allele calls generated from the GenomeStudio software were then corrected using ALCHEMY software (Wright et al., Reference Wright, Tung, Zhao, Reynolds, McCouch and Bustamante2010). Genetic diversity and distances measured 14. Cavalli-Sforza chord (CSChord) (Cavalli-Sforza and Edwards, Reference Cavalli-Sforza and Edwards1967) were estimated using PowerMarker v3.25 (Liu and Muse, Reference Liu and Muse2005). Principal coordinate analysis (PCA) of the Chinese and IRRI samples was performed based on the CSChord (Cavalli-Sforza and Edwards, Reference Cavalli-Sforza and Edwards1967) distance matrix using GENALEX 6 (Peakall and Smouse, Reference Peakall and Smouse2006). The model-based program STRUCTURE (Pritchard et al., Reference Pritchard, Stephens and Donnelly2000) was used to infer population structure and to assign individual varieties into subpopulations. Models with a putative number of subpopulations (K) from 1 to 10 with admixture and correlated allele frequencies were considered (Falush et al., Reference Falush, Stephens and Pritchard2003). Ten independent runs with 20,000 burn-in cycles and 10,000 iterations for each K were implemented based on trial runs of the program. To determine the uppermost level of the K value, Evanno's ΔK was used (Evanno et al., Reference Evanno, Regnaut and Goudet2005). With the same settings of program parameters, STRUCTURE was rerun for assignments of individual Chinese varieties into subgroups based on the maximum of inferred ancestry of individuals. For a graphic view of the parental classification of the Chinese samples, cluster analysis with the 384 SNP markers was carried out using PowerMarker with neighbour-joining tree and viewed in MEGA 5 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).
Results
Genetic diversity of the Chinese and IRRI varieties
Each of the SNP markers, except three showing monomorphism (id3002191, id6010434 and wd5001329), gave two alleles per locus across all of the Chinese and IRRI varieties and resulted in an average of 1.9922 alleles per locus (Table 1). An additional 4 and 37 markers were found monomorphically among the Chinese and IRRI varieties, respectively, when they were analysed separately.
Table 1 Diversity statistics (mean) of the Chinese and IRRI varieties
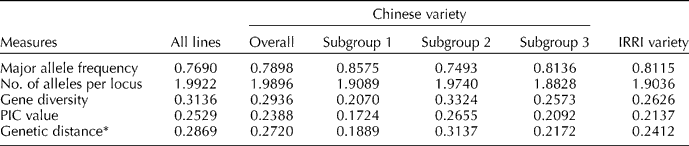
PIC, polymorphism information content.
* Significant differences (P < 0.001) for group means of genetic distances between the Chinese and IRRI varieties, and among the subgroups within the Chinese varieties.
It was observed that the diversity measurements - gene diversity, polymorphism information content (PIC) and genetic distance across all of the samples, were 0.3136, 0.2529 and 0.2869, respectively, which were higher than those indices measured from each individual sample source (CNRRI and IRRI). However, the major allele frequency for all the varieties was 0.7690, which was lower than that estimated from the individual sample sources. Compared with the IRRI varieties, the Chinese samples showed higher values in all of the diversity indices measured except for the major allele frequency. Specifically, the average genetic distance of the Chinese varieties was 0.2720, which was significantly (P < 0.001) higher than the average genetic distance (0.2412) of the IRRI varieties. Both sets of the samples have a minimum of zero distance between pairs, but the maximum distances were 0.5311 and 0.3658, respectively, among the pairs in the Chinese and IRRI variety populations. As for the genetic distance distributions, both populations were skewed to the left, with skewness coefficients of − 0.1300 and − 0.0944 and medians of 0.2817 and 0.2436 for the Chinese and IRRI varieties, respectively (Fig. 1). More pairs (87.3%) of the genetic distances among the IRRI varieties ranged from 0.20 to 0.35 compared with 71.6% of the pairs of the Chinese varieties within that range.
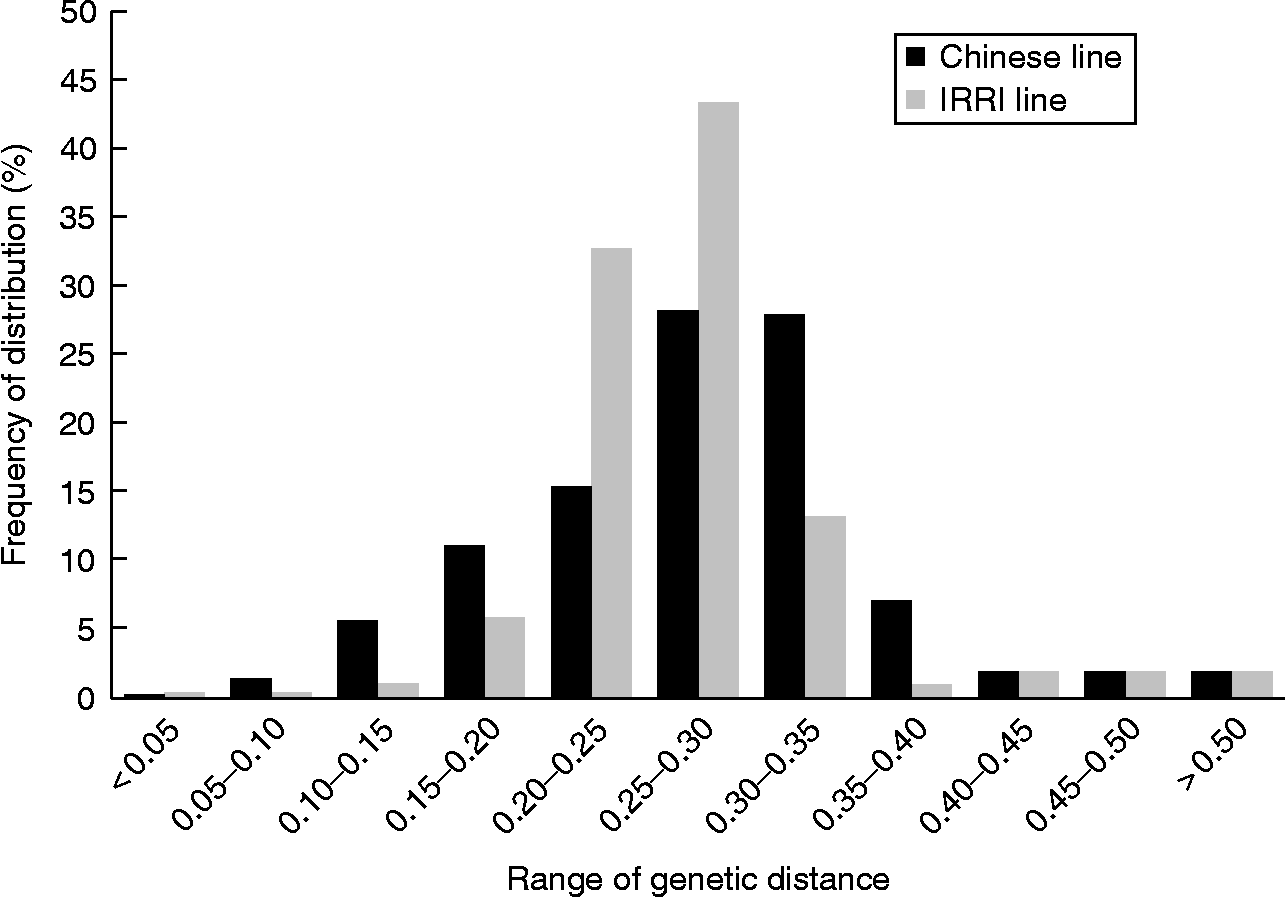
Fig. 1 Distribution of genetic distances of the Chinese and IRRI varieties.
Three pairs among the Chinese varieties (Aijiaonante vs. Xianfeng1hao, Xingheng1hao vs. Jiefang1hao, and Nantehao vs. Zhechang3hao) and two pairs of the IRRI varieties (IR52 vs. IR58 and IRRI146 vs. IRRI150) showed the same SNP genotypes based on RiceOPA2.1 markers.
PCA and population structure
PCA was used to characterize the population structure of the varieties from the two sample sources. The first and second axes of the PCA accounted for 44.5 and 16.0% of the allele variation among populations (Fig. 2). The analysis revealed two clusters among the varieties. All IRRI varieties but one (IR60) were clustered closely together and were clearly separated from the majority of the Chinese varieties. IR60 was an outlier of the IRRI variety group and was closely associated with the majority of the Chinese varieties, specifically close to the varieties from Guangdong province. On the other hand, the Chinese varieties were scattered over a wide range, but most of the varieties from Guangdong were clustered together and showed a relatively close relationship with the IRRI varieties compared with other Chinese varieties from different provinces. However, there was no clear evidence to separate the Chinese varieties by province in the PCA.

Fig. 2 PCA of 215 Chinese and IRRI rice varieties using IRRI SNP RiceOPA2.1 based on CSChord 1967 distance estimates. PC 1 and PC 2 are the first and the second principal coordinates, respectively. Numbers in parentheses refer to the proportion of variance explained by the principal coordinate.
Genetic relationships and differentiation
The two distinct groups (subpopulations) clustered by PCA based on the CSChord genetic distance were verified by the STRUCTURE analysis, which predicted K = 2 as the optimum number of subpopulations (Fig. 3). Individual varieties were assigned into subpopulations based on the maximum population inferred clusters (see Table S1, available online only at http://journals.cambridge.org). The STRUCTURE analysis showed that 94 samples were in subpopulation 1, all composed of the Chinese varieties, and 121 samples in subpopulation 2, which included 67 Chinese varieties and all of the 54 IRRI irrigated inbreds. It was observed again that there was no clear separation to distinguish the individual varieties based on provincial origins among the Chinese varieties. Twenty-one of the 33 varieties from Guangdong were clustered in subpopulation 2, indicating a relatively close association with the IRRI varieties in comparison with varieties from other provinces.
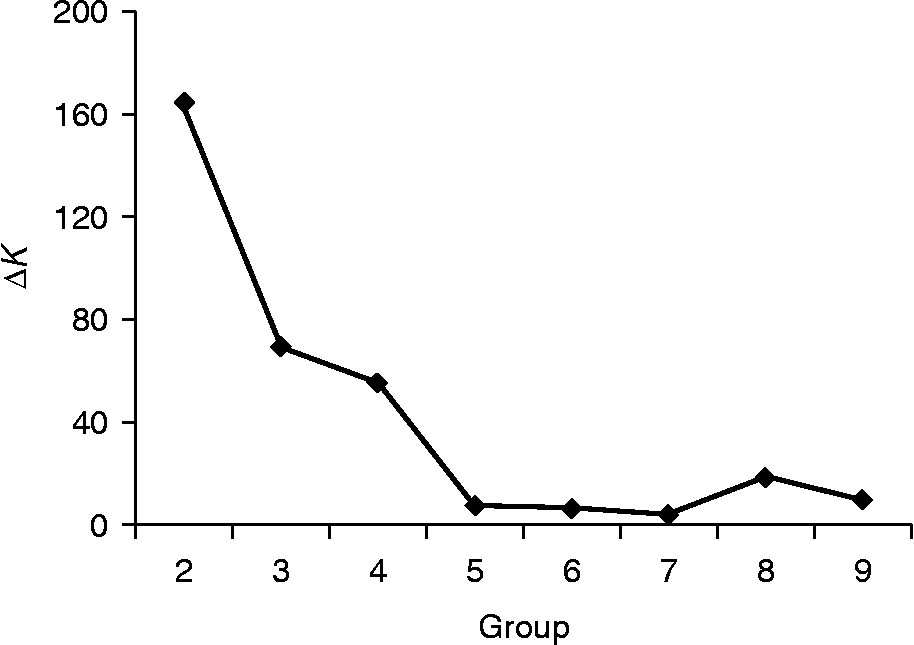
Fig. 3 Likelihood distribution of subpopulations of the Chinese and IRRI varieties based on the average of ΔK over ten runs for each K value.
The Chinese varieties were subclustered into three subgroups with 21, 86 and 54 varieties in subgroup 1, subgroup 2 and subgroup 3, respectively, at the second step of STRUCTURE analysis. It was observed again that the varieties from the different provinces were distributed across the subgroups without clear evidence of separation into subgroups by provinces (see Fig. S1, available online only at http://journals.cambridge.org). The varieties in subgroup 2 were obviously more diverse than the varieties in subgroup 1 or subgroup 3, as indicated in the diversity measurements (Table 1). The average genetic distance of the subgroup 2 varieties was 0.3137, which was significantly (P < 0.001) higher than those measured in subgroup 1 (0.1889) and subgroup 3 (0.2172), showing a divergent population structure in subgroup 2, followed by the varieties in subgroup 3 and subgroup 1 within the Chinese samples.
Discussion
SNP markers
Five rice OPAs were developed at IRRI for the rice SNP platform (Thomson et al., Reference Thomson, Zhao, Wright, McNally, Rey, Tung, Reynolds, Scheffler, Eizenga, McClung, Kim, Ismail, Ocampo, Mojica, Reveche, Dilla-Ermita, Mauleon, Leung, Bustamante and McCouch2012). Our samples were screened using OPA2.1, which was designed for the indica/indica populations. As Thomson et al. (Reference Thomson, Zhao, Wright, McNally, Rey, Tung, Reynolds, Scheffler, Eizenga, McClung, Kim, Ismail, Ocampo, Mojica, Reveche, Dilla-Ermita, Mauleon, Leung, Bustamante and McCouch2012) pointed out, the OPA provides reasonably high polymorphism rates for the targeted germplasm pools, and it could have “an intrinsic bias” affecting the interpretation of diversity results due to the SNP markers being selected with a particular objective in mind, rather than at random. However, the OPA2.1-selected SNP markers are spaced across the genome, informative within and between different germplasm groups, and they have been validated for diversity analysis and fingerprinting of more than 500 rice varieties and breeding lines at IRRI (Thomson et al., Reference Thomson, Zhao, Wright, McNally, Rey, Tung, Reynolds, Scheffler, Eizenga, McClung, Kim, Ismail, Ocampo, Mojica, Reveche, Dilla-Ermita, Mauleon, Leung, Bustamante and McCouch2012). Therefore, we believe that the data generated from this study provide a fair estimation for the genetic diversity for our samples, even though these markers can be further improved for the SNP marker selection of maximum marker efficiency. In this study, 3, 4 and 37 markers showed monomorphism across all of the samples, among the Chinese varieties, and among the IRRI varieties, respectively. The monomorphism of SNP markers in this study is understandable because we used an existing SNP marker package for the diversity analysis. Some markers are not specifically designed and selected for diversity estimation of the population we used. However, those monomorphic markers could be replaced with other markers to provide maximum marker information, especially for those IRRI varieties sharing many ancestral parents in the crosses and that are closely related genetically.
Diversity
It is reasonable to conclude that, among the Chinese and IRRI varieties selected in this study, the Chinese varieties are more divergent than the IRRI varieties based on the marker profiling. These Chinese varieties are from many regions geographically with diverse rice-growing environments, but all of them are indica varieties adapted to the lowland irrigated rice ecosystem. On the other hand, IRRI has been expanding its germplasm sources with more parents involved in variety development. Parents used in irrigated inbred breeding have increased dramatically in the last 20 years (Fig. 4). For example, the pedigree records showed that the number of parents involved in breeding crosses of IR8, IR36, IR64 and IRRI116 (all mega-varieties released in 1966, 1976, 1985 and 1997, respectively), and IRRI154 (released in 2009) were 3, 16, 22, 32 and 43, respectively, showing that the parents in the variety pedigrees increased significantly over the years. However, the IRRI varieties, even though they have been cultivated on more rice area under more diverse environments than the Chinese varieties, are still less diverse than the Chinese varieties, as indicated in this study. We urgently need to diversify IRRI germplasm to be able to adapt to various rice-growing environments and to meet the challenges of climate and rice ecosystem changes. For the hybrid rice breeding programme at the IRRI, specifically, it is more urgent to develop divergent germplasm since the parents and germplasm used in hybrid rice breeding are mostly derived from the IRRI inbred breeding programmes. These sources of limited hybrid rice germplasm are further restrained to only a tiny portion of germplasm available due to the maintaining and restoring requirements. This could be one reason that a lower hybrid heterosis than that of hybrid rice in China is observed in the tropics where IRRI hybrid rice germplasm has been popularly used. With the help of molecular marker tools, hybrid rice germplasm should be regularly monitored and evaluated for genetic composition and population structure to better understand the parental genetic basis and to enhance heterosis.

Fig. 4 Parents involved in variety development of the IRRI irrigated inbred varieties.
All of the Chinese varieties used in this study are improved semi-dwarf HYVs that are widely grown for rice production. They showed a diverse admixture among the groups without a clear separation by province, showing a great degree of genetic integration of alleles. The admixed parents are likely to be the result of shared ancestry in developing varieties due to the widespread use of HYVs. Some of the Chinese varieties, such as those in subgroup 2, are closely allied with the IRRI varieties. This could be due to the sharing of ancestry among those indica varieties with the same semi-dwarf background and a large quantity of IRRI germplasm imported into China, specifically to Guangdong province which has a similar rice ecosystem to the tropical environment, in the last four decades. Germplasm exchange domestically and internationally in China has been playing a very important role in raising the yield potential for local varieties with an accelerated integration of new alleles from different regions and sources, and this greatly enhanced the expansion of the germplasm breeding pools. However, an increased concern is that common HYVs and elite lines/traits are pursued by all of the breeders over the regions, which results in a great similarity of genotypes and phenotypes among the new varieties observed in national and regional rice yield trials. This may result in some unique genes or traits adapted to local environments being lost during line selection under well-controlled selection environments with an increased risk of making the rice ecosystem vulnerable.
Rice heterotic groups
Heterotic hybrids are usually derived between heterotic pools that are defined as those germplasm groups differentiated genetically from each other and can produce superior hybrids when they are crossed. Heterotic pools in rice have not been fully and clearly defined. The concept of rice subspecies or groups clustered based on genetic molecular markers, such as those classified as aus, indica, aromatic, temperate japonica and tropical japonica (Zhao et al., Reference Zhao, Wright, Kimball, Eizenga, McClung, Kovach, Tyagi, Ali, Tung, Reynolds, Bustamante and McCouch2010), is different from the definition of heterotic pools. Heterosis magnitude may be correlated with groups based on molecular markers, but most of the time, the hybrids derived from different subspecies are not applied in commercial production because of various problems with hybrids. However, in general, parental diversity is one of the major contributors to the magnitude of heterosis of hybrid rice, even within a subspecies. The Chinese experience teaches that heterotic rice hybrids are always derived 28 from genetically distance parents. Shanyou 63, a specific hybrid that dominated hybrid rice production in southern and Central China with more than 50% of hybrid rice area coverage in the 1990 s, was bred from the parents of ZS 97 and MH H63. These two lines are also the most widely used parents in China either directly as parents for other hybrids or as crossing donors in parental breeding. ZS 97 is a typical early-season indica variety from southern China and MH 63 is an excellent restorer line derived from a cross involving an IRRI variety (IR30). These two varieties are the representative parents from the two heterotic pools (early-season indica varieties in southern China as female parents and low-latitude indica varieties from IRRI or from other Southeast Asian countries as male parents) identified for three-line hybrid rice in China. However, both of them were clustered into the same subgroup in our study (see Fig. S1 and Table S1, available online only at http://journals.cambridge.org) with a small genetic distance compared with pairs in other different subgroups. It is noteworthy that many commercial hybrids have been produced from ZS 97, but no heterotic hybrids have been found or commercialized in varieties associated closely with ZS 97, i.e. some hybrid rice parents derived from ZS 97 crosses have not produced heterotic hybrids as good as ZS 97 itself. Some alleles of japonica rice were found in ZS 97 (McNally et al., Reference McNally, Childs, Bohnert, Davidson, Zhao, Ulat, Zeller, Clark, Hoen, Bureau, Stokowski, Ballinger, Frazer, Cox, Padhukasahasram, Bustamante, Weigel, Mackill, Bruskiewich, Ratsch, Buell, Leung and Leach2009). It is not clear what role those japonica alleles played for the combining ability of heterotic hybrids and what genetic factors contributed molecularly to the heterosis of ZS 97 hybrids. Molecular profiling of those varieties associated closely with ZS 97 and MH 63 and further hybridization studies may provide some hints of understanding the basis of hybrid heterosis in hybrid rice and to further enhance heterosis in the three-line hybrids as well as to serve as a guideline reference for developing two-line hybrid rice. Hybrid heterosis has not been fully explored among the parents from our parental groups based on molecular markers. Heterotic pools in hybrid rice could be different from the parental groups clustered by molecular markers. However, information from molecular markers with field studies of heterotic pools will offer great help in developing heterotic hybrids by selecting parents from different heterotic pools or specific molecular combinations. Parental classification with assignment of populations and individual varieties into groups would help to develop heterotic rice hybrid and provide a basis for parental selection based on heterotic pools.
Acknowledgments
This research was supported by the Hybrid Rice Development Consortium at IRRI and a project of IRRI-NSFC (National Nature Science Foundation, China, grant no. 30921140408). We thank Dr Michael Thomson and his team in the Molecular Marker Applications Laboratory at the IRRI for genotyping the samples.







