INTRODUCTION
Parasitic nematodes of the superfamily Thelastomatoidea live in the hindgut of saprophytic terrestrial arthropods. They are transmitted directly via an infective egg, and have a haplo-diploid life-cycle in which haploid males arise from unfertilized eggs and females arise from fertilized eggs (Adamson, 1989). Simultaneous infections with multiple species, called ‘parasite guilds’, are common for the Thelastomatoidea and have been the subject of numerous studies examining the forces that structure them (Hominick and Davey, 1972; Zervos, 1988; Adamson and Noble, 1992, 1993; Morand and Rivault, 1992; Connor and Adamson, 1998; Müller-Graf et al. 2001).
The accurate identification and genetic characterization of these nematodes, irrespective of developmental stage and sex, is central to studying their systematics (i.e., taxonomy and phylogeny), population genetics and ecology. Individual adult thelastomatoids are typically identified and distinguished on the basis of morphological features, which include dimensions of the oesophagus, position of nerve ring and excretory pore, shape and orientation of the reproductive systems in females and males, number and orientation of copulatory papillae in males, and tail shape (Cobb, 1920; Skryabin et al. 1984; Adamson and van Waerebeke, 1992a,b,c). Although using these morphological characters, females can be identified to species, this is not always possible for males. Also, it is not possible to use these characters to specifically identify eggs or larvae to species. In addition, the identification of these parasites to species is hampered by the lack of information about their host specificity and geographical distribution.
Currently, there are no DNA sequence data for the Thelastomatoidea in current gene databases and only few are available for the other Oxyurida species. For example, sequences of the small subunit (SSU) of the nuclear ribosomal RNA gene were published for species of Dentostomella (Nematoda: Oxyuroidea) (Blaxter et al. 2000) and for the human pinworm Enterobius vermicularis (see Iniguez et al. 2002, 2003). However, little is known about the utility of ribosomal gene loci for the purposes of specific identification and/or for determining genetic relationships within the Oxyurida in general.
Recently, the complete sequence of the large subunit of the nuclear ribosomal RNA gene (LSU) for Labiostrongylus bipapillosus (Nematoda: Strongyloididae), a parasite of the eastern grey kangaroo, Macropus giganteus was published (Chilton et al. 2003), which was the first report of a complete LSU sequence for any parasitic nematode. Comparison of this gene sequence with that of the free-living nematode Caenorhabditis elegans identified 12 variable expansion segments, called divergent (D) domains (D1–D12). These domains differed significantly in length and sequence between the two nematodes. In a more recent study, one of these 12 domains (D3 domain) was demonstrated to be useful for the genetic characterization of species and/or genotypes of Trichinella (class Adenophora; Trichinellida) (Gasser et al. 2004), which suggested that the D3 domain should be applicable to other nematode groups, including the Thelastomatoidea.
The aims of the present study were to sequence the D3 domain for 5 species of thelastomatoid from cockroaches from the eastern coast of Australia, to examine the magnitudes of sequence variation in this domain within each of and among 5 morphospecies, and to explore the genetic relationships of these species.
MATERIALS AND METHODS
Adult female specimens of Aoruroides queenslandensisJex, Cribb and Schneider, 2004 (n=3), Blattophila sphaerolaimaCobb, 1920 (n=1), Cordonicola gibsoni Jex, Schneider, Rose and Cribb, 2005 (n=9), Desmicola ornata Jex, Schneider, Rose and Cribb, 2006 (n=8), and Leidynemella fusiformis Cobb in Chitwood and Chitwood, 1934 (n=3) were collected from Panesthia cribrata Saussure (n=2), a wood burrowing cockroach, from Beerburrum State Forest, Queensland, Australia (28 °58′S, 152 °58′E). The nematodes were identified morphologically to species, as described recently (Jex et al. 2006).
Total genomic DNA was released from individual adult specimens by suspending each of them in 10 μl of H20 and 5 μl of 1 M Tris (pH 7·6), incubation at 92 °C for 10 min, cooling on ice for 5 min, addition of 1 μl of proteinase K (20 mg/ml), incubation at 48 °C for 6 h, inactivation of the enzyme at 92 °C for 10 min and centrifugation at 10000 g for 5 min. The supernatant fraction containing DNA was used immediately or frozen at −20 °C until use. Genomic DNA from Oxyuris equi (Nematoda: Oxyurida) was isolated and purified as described previously (Hu et al. 2002).
The D3 domain of the LSU was amplified by the PCR using primers NC28-26 (5′-ACCCGTCTTGAAACACGGA-3′) and NC28-25R (5′-GATTAGTCTTTCGCCCCTA-3′) (Chilton et al. 2003). In brief, 5 μl of genomic DNA suspension were added directly to the PCR mix (50 μl; overlaid with paraffin oil) containing 250 μM of each dNTP, 4·0 mM MgCl2, 25 pmol of each primer and 2 U of Taq polymerase (Promega), placed immediately on a freeze block (−20 °C) and then subjected to cycling in a 480 thermal cycler (Perkin Elmer Cetus) at 94 °C, 5 min (initial denaturation), followed by 40 cycles of 94 °C, 1 min (denaturation), 55 °C, 1 min (annealing) and 72 °C, 1 min, followed by 72 °C for 5 min. Samples without DNA (no-DNA) or with cockroach (host) DNA were included as control samples.
Amplicons were purified over mini-columns (Wizard™ PCR Prep, Promega, WI, USA), eluted in 30 μl of H2O and then subjected to automated sequencing (version 3.1; BigDye® chemistry, Applied Biosystems) using the same primers as for the PCR. The sequences have been deposited in gene databases. The sequences were verified (in relation to the electropherograms) and aligned manually. Sequences were compared (using BLAST, available at http://www.ncbi.nlm.nih.gov/BLAST/) with sequences from other taxa lodged in the GenBank database (i.e., partial 28S rRNA genes available for Ascaris suum and Necator americanus (see Nadler and Hudspeth, 1998; Nadler et al. 2000)) and aligned using the program ClustalX (Thompson et al. 1997). Pairwise comparisons of sequence differences (D) were made using the formula D=1−(M/L) (Chilton et al. 1995), where M is the number of alignment positions at which the two sequences have a base in common, and L is the total number of alignment positions over which the two sequences are compared. Phylogenetic analyses of the nucleotide sequence data for the D3 domain were conducted in PAUP* 4.0b10 (Swofford, 1999). As there is no consensus opinion as to which is the ‘optimum’ approach for phylogenetic reconstruction, different methods were employed, and the congruence among the trees produced was established. The ‘exhaustive’ heuristic neighbour-joining method, employing the LogDet distance model, was used to construct trees from distance data, and a consensus tree was generated. The maximum likelihood and maximum parsimony methods (based on character state analysis) were also employed; heuristic searches were performed with tree-bisection reconnection (TBR) branch-swapping, alignment gaps were treated as missing data, and bootstrap analyses (1000 replications) were conducted using the MulTrees option. The inference model used for the maximum likelihood analysis was the Kimura two-parameter model. The outgroup employed was Oxyuris equi, a member of superfamily Oxyuroidea, the only known sister superfamiliy to the Thelastomatoidea.
RESULTS
The D3 domain was amplified from 24 individual specimens representing 5 species of Thelastomatoidea, and 1 species of Oxyuroidea, Oxyuris equi. On agarose gels, the amplicons produced varied in size from 300–340 bp, and no size variation was detected within each of the 4 morphospecies for which multiple samples were examined. All 24 amplicons were sequenced, and the sequences obtained varied from 275–282 bp in length and had a G+C content of 51–53%.
No sequence variation was detected within Desmicola ornata (n=7; D3 sequence length: 281 bp), whereas sequence variations of 0·4% (1 bp), 0·4–1·4% (1–4 bp) and 1·8% (5 nt) were detected within Aoruroides queenslandensis (n=3; 282 bp), Cordonicola gibsoni (n=9; 279–281 bp) and Leidynemella fusiformis (n=3; 279 bp), respectively. The sequence for 1 specimen of Blattophila sphaerolaima was 275 bp in length. The alignment of the sequences representing all 5 species is shown in Fig. 1. Upon pairwise comparison, the 5 thelastomatoid species differed in the D3 sequence from Oxyuris equi by 11·7–17·6% (29–47 bp). Within the Thelastomatoidea, Aoruroides queenslandensis differed in sequence by 4·4–14·5% (12–40 bp), B. sphaerolaima by 9·1–12·4% (25–34 bp), C. gibsoni by 5·0–12·4% (12–31 bp), D. ornata by 4·3–9·6% (12–30 bp) and L. fusiformis by 7·4–12·4% (28–40 bp) from the heterologous species of thelastomatoids (Table 1).

Fig. 1. Alignment of the D3 domain sequences for species of Thelastomatoidea, Aoruroides queenslandensis (Aq1, 2), Blattophila sphaerolaima (Bs1), Cordonicola gibsoni (Cg1-Cg3), Desmicola ornata (Do1), Leidynemella fusiformis (Lf1–Lf3) and Oxyuroidea, Oxyuris equi (Oe). Differences indicated with an asterisk (*). An alignment gap is shown as a hyphen (-). Vertical slash (|) represents every tenth base pair of the sequence. Number at the end of the top line represents every 60th base pair.
Table 1. Pairwise differences (%) in the D3 sequence within and among 5 thelastomatoid morphospecies, Aoruroides queenslandensis, Blattophila sphaerolaima, Cordonicola gibsoni, Desmicola ornata and Leidynemella fusiformis, and one oxyuroid, Oxyuris equi (n, The number of individuals sequenced per species.)

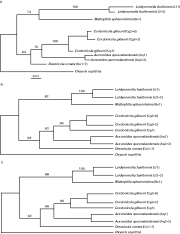
Phylogenetic analysis of the D3 sequence data was performed using the 3 different tree-building methods. The trees produced using these methods had a similar topology and bootstrap support (see Fig. 2). Individual species of thelastomatoid resolved separately from one another. Bootstrap support for the groups of individual species ranged from 70 to 100% for the neighbour-joining method, 62 to 100% for the maximum parsimony method and 66 to 100% for the maximum likelihood method. All 3 tree-building methods produced trees which were divided into 2 major groups; one comprised L. fusiformis and B. sphaerolaima (73–88% bootstrap support) and a second included A. queenslandensis, C. gibsoni and D. ornata (62–64% bootstrap support). Within the group formed by L. fusiformis and B. sphaerolaima, the bootstrap support for the separate resolution of L. fusiformis from B. sphaerolaima was 100% using all 3 tree-building methods. Within the A. queenslandensis/C. gibsoni/D. ornata-group, a subgroup including A. queenslandensis and C. gibsoni was resolved, with bootstrap support of 62–70%. C. gibsoni and A. queenslandensis grouped separately, supported by a bootstrap value of 90–100%.

Fig. 2. Phylogenetic analysis of the D3 domain sequence data for Aoruroides queenslandensis, Blattophila sphaerolaima, Cordonicola gibsoni, Desmicola ornata and Leidynemella fusiformis, with Oxyuris equi as the outgroup, using 3 tree-building methods: neighbour-joining, maximum parsimony and maximum likelihood methods (panels A, B and C, respectively). Bootstrap support values are indicated. Sequence codes corresponding to each node on the three trees presented are shown in enclosed parentheses and are consistent with the sequence codes used in Fig. 1.
DISCUSSION
Morphological studies have led to a large body of knowledge for the Thelastomatoidea (Cobb, 1920; Leibersperger, 1960; Skryabin et al. 1984; Adamson and van Waerebeke, 1992a,b,c). However, there are significant limitations in a taxonomy based on morphology alone. The descriptions of many of the taxa within the Thelastomatoidea are dated, and there has been little consensus in the characters used or the format employed when describing new taxa (Adamson and van Waerebeke, 1992a). Subjective character states, such as ‘long’, ‘short’, or ‘medium’, are commonly used but are problematic, as they are not objectively defined. Additionally, male thelastomatoids are often difficult to identify due to a lack of informative morphological characters and dissimilarity in the morphology of males and females of the species. As multiple species guilds are common in thelastomatoid infections, matching males to females is difficult, thus impeding the use of male thelastomatoids to discern taxonomic relationships. These potential difficulties can lead to uncertainty in the identification of thelastomatoids. An objective and robust method for identifying these parasites is necessary.
In the present study, we examined sequence variation in the D3 domain of the LSU rRNA gene from multiple specimens representing 5 thelastomatoid species that parasitize Australian burrowing cockroaches. The identification of these species has been based upon examination of morphological characters in relation to previous descriptions (Jex et al. 2004, 2006). The current morphology-based taxonomy for the group suggests that the 5 species examined here represent 5 different genera within one family, the Thelastomatidae. Analysis of the D3 domain data in the present study revealed that the sequence variation detected within species (0–1·8%), and interpreted to represent population variation, was considerably lower than the differences between species (4·3–12·4%). Three of the thelastomatoid species examined herein, A. queenslandensis, C. gibsoni and D. ornata, had interspecific sequence differences of 4·3–6·7% (not including B. sphaerolaima or L. fusiformis). The remaining 2 species, B. sphaerolaima and L. fusiformis, had higher levels of genetic difference (7·4–12·4% and 8·5–12·1% respectively).
Phylogenetic analyses of the D3 domain data set examined here have several implications. Within the Thelastomatoidea, each of the 5 morphospecies formed monophyletic clades, irrespective of the tree-building method applied (bootstrap support: 62–100%), demonstrating that the D3 domain provides sufficient information to resolve thelastomatoids at the genus level. Bootstrap support for the resolution of D. ornata, with respect to A. queenslandensis and C. gibsoni, was somewhat low (62–66%); however, as the D3 sequences for all 8 D. ornata were the same, there is confidence that the division is legitimate. In addition to the divisions at the species level, 2 major clades were formed by the species analysed in this study; one formed by L. fusiformis and B. sphaerolaima, and a second formed by A. queenslandensis, C. gibsoni and D. ornata. The current taxonomic hypothesis for the Thelastomatoidea divides the superfamily into 5 families, the Hystrignathidae, Protrelloididae, Pseudonymidae, Thelastomatidae and Travassosinematidae (Adamson, 1989). Adamson (1989) found morphological characters to justify the monophyletic lineage of all of these families except the Thelastomatidae and suggested that the latter was probably paraphyletic. However, as this author could find no legitimate basis for splitting the family, he retained it. The thelastomatoid species studied herein are all currently considered to be within the family Thelastomatidae. The present results appear to suggest the resolution of at least 2 monophyletic lineages within the Thelastomatidae, which may provide the future basis for possible division of this family. However, clearly, further analysis of numerous species within the family is required before any firm conclusion can be made.
The D3 domain may be suitable as a genetic marker for exploring the ecology of the thelastomatoids, such as the extent of geographical ranges and levels of host specificity. This is particularly important, given that many of the thelastomatoid species examined herein have been reported from hosts distributed across much of eastern Australia, of which as many as 33 species are burrowing cockroaches (Jex et al. 2004, 2006). Additionally, thelastomatoid specimens taken from panesthiine cockroaches from Thailand and Indonesia appear to be morphologically the same as several of the species studied in Australia (Aaron Jex, unpublished findings). Furthermore, there is morphological evidence to suggest that several species, including Cordonicola gibsoni, Desmicola ornata and Leidynemella fusiformis, parasitizing panesthiine cockroaches in fallen logs in the Lamington National Park, Queensland, Australia, also parasitize wood-feeding millipedes (Diplopoda: Polydesmidae and Glomeridae) and beetles (Coleoptera: Passalidae) in the same logs (Aaron Jex, unpublished findings). These observations indicate that the host specificity of many of these parasites is low and that the geographical distributions are vast. However, given the large range of hosts and geographical regions reported for these species, the potential for the presence of cryptic species is great. Molecular methods provide a means of complementing morphological approaches of identification. Thus, the genetic markers in the D3 domain may be useful to test the validity of the morphological identification of these species and aid in the determination of the extent to which Australian thelastomatoids are shared among arthropods within and outside of Australia. Such analyses would increase our knowledge of the historical, biogeography and evolutionary lineages of these parasites. However, in order to utilize this approach, the levels of intraspecific variation and interspecific differences in the D3 must be rigorously established and validated for a wide range of species of thelastomatoid. Based on the present data set, interspecific differences and intraspecific variation are estimated to be 1·5–4·5% and 0·0–1·5%, respectively. However, the individual species studied herein represent separate genera and, thus, these estimates are uncertain. Hence, further detailed analysis of many congeneric thelastomatoid species groups (from broader host and geographical ranges) is needed to establish the levels of intraspecific variation and interspecific differences in the D3 domain sequence. If the degree of sequence variation in the D3 domain within well-defined morphospecies remains low, it provides the prospect for the detection of cryptic species using the D3 domain together with mitochondrial loci, such as the nicotinamide dehydrogenase subunit 4 (nad4) and cytochrome c oxidase subunit 1 (cox1) genes. The latter mitochondrial genes have been shown to be particularly well suited for this purpose within the Nematoda (reviewed by Blouin, 2002).
In conclusion, this study represents the first molecular characterization of members of the Thelastomatoidea. We conclude that the D3 domain of the LSU is useful in determining genetic relationships of thelastomatoids and propose its use in studies of the systematics and ecology of this nematode group.
This study was supported by the Queen Elizabeth II Centennial Scholarship (Government of British Columbia, Canada), the University of Queensland International Postgraduate Research Scholarship, the International Postgraduate Research Scholarship (Government of Australia), the Australian Biological Research Study, the Australian Academy of Science and Australian Research Council. The authors thank Darron Moates and Scott O'Neill from the Beerburrum Forestry division of the Department of Primary Industries, Queensland, Australia and Ian Bryant and Leigh Klienschmidt from Queensland Parks and Wildlife, Queensland, Australia, for access to sampling sites and provision of material and processing of collection permits. Thanks are also due to Dr Youssef Abs EL-Osta for advice on genomic DNA extraction and the PCR, and to 2 anonymous reviewers for their constructive comments on the manuscript.