Introduction
Feeding during early life stages is considered one of the main factors driving year class strength of fish stocks (Paulsen et al., Reference Paulsen, Clemmesen and Malzahn2014), being mostly studied by means of the traditional ‘gut content’ method. However, this approach cannot easily discriminate between organisms that are either assimilated, egested or pass through the gut undigested (Pitt et al., Reference Pitt, Connolly and Meziane2009). Besides, it can underestimate the importance of soft and highly digestible items and overestimate that of recently consumed items (Graeve et al., Reference Graeve, Dauby and Scailteur2001), providing only a snapshot of the recent diet. In this context, the ‘fatty acid trophic markers approach’ (FATM) appears as a potential tool for qualitatively assessing predator diets and prey assimilation (Dalsgaard et al., Reference Dalsgaard, St. John, Müller-Navarra and Hagen2003; Budge et al., Reference Budge, Iverson and Koopman2006; Iverson, Reference Iverson, Arts, Brett and Kainz2009). The basis of FATM is that a consumer incorporates the ‘marker’ or ‘signature’ of its food source into its somatic tissue with minimal or predictable change, thus providing an integrated record of dietary intake over time.
Fatty acids (FA) represent the building blocks of lipids found in all organisms. Saturated (SFA) and monounsaturated (MUFA) FA can be biosynthesized de novo by most fish (reviewed by Dalsgaard et al., Reference Dalsgaard, St. John, Müller-Navarra and Hagen2003), while polyunsaturated fatty acids (PUFA) usually cannot. The latter are first synthesized by primary producers and then incorporated unchanged into the tissues of secondary consumers. FA are particularly useful in that they are relatively easily measured, integrative, sensitive and responsive to change in a predictable manner (Iverson, Reference Iverson, Arts, Brett and Kainz2009). In this sense, some FA have a unique origin in certain taxa, thereby allowing these groups to be distinguished (Pitt et al., Reference Pitt, Connolly and Meziane2009). For example, widely used are the bacterial FA markers 15:0, 17:0 and 18:1n-7 (Volkman et al., Reference Volkman, Johns, Gillian and Perry1980; Kaneda, Reference Kaneda1991); phytoplankton FA markers for diatoms (16:1n-7 and ratio 16:1/16:0 > 1) and dinoflagellates (18:4n-3, 22:6n-3) (St. John & Lund, Reference St. John and Lund1996; Mansour et al., Reference Mansour, Volkman, Jackson and Blackburn1999); FA synthesized by marine calanoid copepods such as 20:1 and 22:1 (Sargent & Falk-Petersen, Reference Sargent and Falk-Petersen1988); or the ratio 18:1n-9/18:1n-7 > 1 indicating a carnivorous diet (e.g. Hagen et al., Reference Hagen, Kattner, Terbruggen and Van Vleet2001; Nelson et al., Reference Nelson, Mooney, Nichols and Phleger2001; Phillips et al., Reference Phillips, Nichols and Jackson2003).
Argentine hake Merluccius hubbsi Marini 1933 supports the major demersal finfish fishery in the Argentinean Continental Shelf (ACS). Of two main stocks, the southern or Patagonian one (between 41°–55°S) is the most abundant, accounting for 85% of total hake biomass in the ACS (Aubone et al., Reference Aubone, Bezzi, Castrucci, Dato, Ibáñez, Irusta, Pérez, Renzi, Santos, Scarlato, Simonazzi, Tringali, Villarino, Bezzi, Akselman and Boschi2000). This stock spawns during late spring (December) and summer (January–March) in the north Patagonian shelf (NPS) between 43°–45°30′S (Pájaro et al., Reference Pájaro, Macchi and Martos2005). In December, spawning is located near Isla Escondida (>50 m in depth), but throughout the summer it extends to the east and south reaching the Bahía Camarones area, where maximum annual densities of eggs and larvae (<20 mm total length) have been reported (Ehrlich & Ciechomski, Reference Ehrlich and Ciechomski1994). During summer months, larvae undergo a weak onshore and south-westward drift, persisting in the main spawning ground between Isla Escondida and Bahía Camarones due to retention mechanisms (Álvarez Colombo et al., Reference Álvarez Colombo, Dato, Macchi, Palma, Machinandiarena, Christiansen, Betti, Derisio, Martos, Castro-Machado, Brown, Ehrlich, Mianzan and Acha2011). Within this nursery scenario, Temperoni & Viñas (Reference Temperoni and Viñas2013) – based on traditional gut content analyses – determined that hake larvae are specialist predators upon copepodites and adults of calanoid copepod species such as Drepanopus forcipatus and Calanoides carinatus, in agreement with feeding preferences of other Merluccius species (e.g. Morote et al., Reference Morote, Olivar, Bozzano, Villate and Uriarte2011 and references therein). Despite the growing body of fatty acid-related data pertaining to fish larvae and zooplankton worldwide, the FA approach has not been applied yet in the ACS to depict M. hubbsi larval feeding preferences. Therefore, this study aims to (1) provide the first data on fatty acid profiles of M. hubbsi larvae and their copepod prey, and (2) either confirm or broaden the diet information gathered from the traditional analyses. Results are expected to provide new insight into hake larvae trophic ecology, and should be useful in the context of M. hubbsi ongoing recruitment studies.
Materials and methods
Sampling
Merluccius hubbsi larvae and their copepod prey were collected in 11 sampling stations during two research surveys carried out by the Instituto Nacional de Investigación y Desarrollo Pesquero (INIDEP) on board the RV ‘Eduardo Holmberg’ in the NPS in January 2012 (N = 4) and 2013 (N = 7) (Figure 1). Larvae were captured during daylight with a 300 µm-mesh Bongo net, while copepods were obtained with a 200 µm-meshed Minibongo net in the same stations. Tows were oblique from near the bottom to the surface, with bottom depths ranging from 49.0 to 99.0 m. Immediately after collection, hake larvae and copepods were rinsed with distilled water, blotted dry on tissue paper and stored separately in cryovials in liquid nitrogen at −70°C. Once in the laboratory, samples were stored at −80°C until extraction.

Fig. 1. Sampling stations in the north Patagonian shelf where Merluccius hubbsi larvae and copepods were collected during January 2012 (squares) and 2013 (circles).
Laboratory analyses
Hake larvae (~N = 10 larvae per station) and copepods (~N = 100 copepods from each species) were pooled in each sampling station (N = 11) to generate sufficient material for an adequate signal. Mean total length (±SD) of larvae was 6.2 (1.2) mm (larvae measured: N = 29) and 6.3 (1.9) mm (N = 140) in January 2012 and 2013, respectively. An inspection of the copepod samples under stereomicroscope showed dominance of copepodites and adults of D. forcipatus and C. carinatus. Both species, known as main prey of hake larvae, were analysed together under the ‘copepods’ taxonomic heading, due to their similarity in terms of FA profiles (Falk-Petersen et al., Reference Falk-Petersen, Sargent, Lønne and Timofeev1999; Cripps & Atkinson, Reference Cripps and Atkinson2000) and feeding preferences (Antacli, Reference Antacli2011; B. Santos, personal communication). After manually processing the samples with a Potter–Elvehjem tissue homogenizer, total lipids were extracted by Bligh & Dyer (Reference Bligh and Dyer1959), and FA were determined using gas chromatography (GC) following transesterification to FA methylesters (FAME). Procedures for FA methylation were based on ISO 12966-2 (International Organization for Standardization, 2017), with some modifications. Briefly, 60 mg of lipid sample were mixed with 2 ml hexane and 0.3 ml of KOH/MeOH reagent in a glass tube. The sample was mixed vigorously (1 min) with a vortex. Then, 2 ml of NaCl and 2 ml of hexane were added and mixed again for 1 min. The sample was allowed to stand for 5 min, and the upper hexane layer was separated and transferred to a clean tube. FAME were determined with a Shimadzu GC-2010 (Shimadzu Corp., Kyoto, Japan), equipped with a flame-ionization detector (260°C) and capillary column (30 m × 0.32 mm; 0.25 µm film thickness; Omegawax 320). GC parameters were set as follows: split rate 50, injector temperature 250°C, column temperature 120°C and nitrogen as a carrier gas. The oven temperature was increased to 240°C at a rate of 5°C min−1 and held for 5 min. A volume of 1 µl of sample was manually injected and FA peaks were identified by comparison of their retention times with those of external reference standards (Supelco FAME Mix C4-C24 + PUFA No 1 Marine Source). Individual FA data were reported as % peak area of the total FAME area.
Data analyses
Fatty acids representing ≥1% of the total as well as those usually known as potential tracers of food items were analysed using multivariate routines in the statistical software package PRIMER 6.1.13 (Clarke & Gorley, Reference Clarke and Gorley2006) with PERMANOVA + 1.0.3 (Anderson et al., Reference Anderson, Gorley and Clarke2008). The data were left untransformed following Cook et al. (Reference Cook, Shucksmith, Orr, Ashton and Berge2010) to prevent an excessive weighting to fatty acids with a low contribution to the profiles. Permutational multivariate analysis of variance (PERMANOVA) performed on Bray–Curtis similarity matrix was used to assess differences between fatty acids composition based on sampling years (two levels: 2012 and 2013; fixed) and items (two levels: larvae and copepods; fixed). The significance of PERMANOVA analysis (set at P < 0.01) was determined using permutation of residuals under a reduced model (4999 permutations) with type III sums of squares (Anderson et al., Reference Anderson, Gorley and Clarke2008). The similarity percentages (SIMPER) routine was used to identify fatty acids that contributed most to observed differences in hake larvae and copepods profiles. Data were visualized with multidimensional scaling (MDS), and the stress value represented the goodness of fit for the ordination. Stress value <0.2 was considered to be acceptable, while plots with stress values >0.2 are close to random (Clarke & Gorley, Reference Clarke and Gorley2006). To aid in data interpretation, fatty acids were represented in the MDS with vectors of relative length corresponding to their strength (i.e. magnitude of change and variability) in sample positioning. Correlation coefficients (Pearson's r) of the fatty acids and the MDS 1 and MDS 2 were also calculated.
Additionally, and to judge the significance of the similarity between hake larvae and copepods profiles, fatty acids compositional data of other potential prey items were collected from the literature, since no real samples or previous studies were available for the north Patagonian shelf. Available data for the Argentinean Continental Shelf corresponded to fatty acid profiles of diatoms and the calanoid Acartia tonsa obtained in a human-impacted estuary during spring (Bahía Blanca estuary, 38°45′–39°40′S 61°45′–62°30′W) (Dutto et al., Reference Dutto, Kopprio, Hoffmeyer, Alonso, Graeve and Kattner2014), as well as of the euphausiid E. lucens collected south of the study area in January (San Jorge Gulf, 45°–47°S 65°W) (Temperoni, Reference Temperoni2015). In addition, and considering the dominant protozoan genera reported during austral summer in the north Patagonian shelf (Carreto et al., Reference Carreto, Carignan, Montoya, Cucchi Colleoni, Sánchez and Bezzi2007), profiles of the dinoflagellates Heterocapsa, Alexandrium, Gymnodinium and Prorocentrum (Hallegraeff et al., Reference Hallegraeff, Nichols, Volkman, Blackburn and Everitt1991; Mansour et al., Reference Mansour, Volkman, Jackson and Blackburn1999, Reference Mansour, Frampton, Nichols, Volkman and Blackburn2005; Dijkman & Kromkamp, Reference Dijkman and Kromkamp2006; Hammann et al., Reference Hammann, Tillmann, Schröder and Vetter2013) as well as of the ciliate Strombidium (Broglio et al., Reference Broglio, Jónasdóttir, Calbet, Jakobsen and Saiz2003) were included. The same multivariate analyses previously described were used to inspect similarity among profiles of hake larvae and these potential prey.
Results
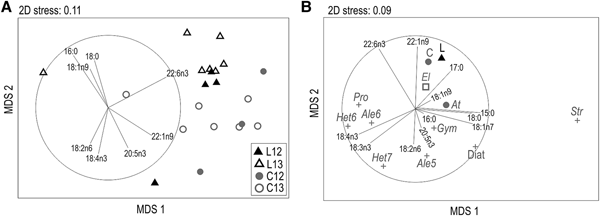
PERMANOVA results showed no significant differences between years of sampling (Pseudo-F = 1.25, P[perm] = 0.28) in the fatty acids profiles of hake larvae and their copepod prey (Table 1). On the contrary, a significant difference was observed in composition between both groups (Pseudo-F = 3.93, P[perm] = 0.01). Major FA of hake larvae and copepods were the SFA 16:0 and 18:0, the MUFAs 18:1n-9 and 22:1n-9, and the PUFA 22:6n-3. These fatty acids contributed to 86.11 and 84.95% of the similarity (SIMPER test, Table 2) in larvae and copepods profiles, respectively (Table 2). Although in lower percentages, typical FA markers of bacteria (i.e. 15:0, 17:0, 18:1n-7) and dinoflagellates (18:4n-3, 22:6n-3) were observed in larvae and copepods profiles, as well as a FA ratio indicative of a more carnivorous diet >1 (18:1n-9/18:1n-7) in the larvae. MDS revealed a differentiation in the spatial ordination of hake larvae and copepod samples with a moderate goodness of fit (stress value: 0.11; Figure 2A). The FA with the highest effect on the sample ordination, based on Pearson's correlation, were 22:6n-3 and 22:1n-9 in the MDS 1 and 16:0, 18:0 and 18:4n-3 in the MDS 2 (Table 3a). 22:6n-3 and 16:0 were the fatty acids that contributed most to similarity within as well as to dissimilarity between larvae and copepods groups. Higher proportions of these fatty acids together with 18:1n-9 were observed in larvae, while copepods samples showed a higher contribution of 22:1n-9 and 18:4n-3.

Fig. 2. MDS ordination of (A) Merluccius hubbsi larvae (L) and copepods (C) fatty acid profiles sampled during January 2012 (12) and 2013 (13) in the north Patagonian shelf, (B) M. hubbsi larvae (L) and copepods (C) fatty acids profiles of this study and other potential prey items profiles collected from the literature (for abbreviations see Table 4).
Table 1. Fatty acids mean percentage (standard deviation) of total FAME in Merluccius hubbsi larvae and copepods during January 2012 and 2013 in the north Patagonian shelf

nd, not detected.
Table 2. Similarity percentages (SIMPER) analysis showing major fatty acids contributors to the average similarity within and to the average dissimilarity between both groups (Merluccius hubbsi larvae and copepods)

Table 3. Correlation coefficients (Pearson's r) of the fatty acids and the dimensions (MDS 1 and 2) in the MDS ordinations including fatty acids profiles from (a) Merluccius hubbsi larvae and copepods of this study, and (b) M. hubbsi larvae and copepods from this study as well as potential prey items collected from the literature

Only fatty acids strongly correlated (r ≥ 0.6) with MDS 1 or MDS 2 are presented.
When comparing hake larvae profiles with those from other potential prey items collected from the literature (Table 4), MDS showed a moderate goodness of fit (stress value: 0.09; Figure 2B). Larvae clustered together with copepods from the study area at 85% similarity (CLUSTER not shown). Euphausia lucens profiles arranged close to larvae and copepods, mainly due to high percentages of 22:1n-9 and 22:6n-3 along MDS 2 (Table 3b). Dinoflagellates from genera Heterocapsa, Prorocentrum and Alexandrium arranged close together along MDS 1, and their profiles were characterized by a high contribution of 18:4n-3 and 18:3n-3. On the opposite side of the MDS 1, diatoms and the ciliate Strombidium had a higher contribution of 18:1n-7 and 15:0.
Table 4. Collection of mean fatty acids (FA) profiles (% of total) from the literature to compare similarity of M. hubbsi larvae profiles to copepods in the north Patagonian shelf over other potential prey species option

nd, not detected; tr, trace.
Data sources: (1) Dutto et al. (Reference Dutto, Kopprio, Hoffmeyer, Alonso, Graeve and Kattner2014); (2) Broglio et al. (Reference Broglio, Jónasdóttir, Calbet, Jakobsen and Saiz2003); (3) Hallegraeff et al. (Reference Hallegraeff, Nichols, Volkman, Blackburn and Everitt1991); (4) Mansour et al. (Reference Mansour, Volkman, Jackson and Blackburn1999); (5) Hammann et al. (Reference Hammann, Tillmann, Schröder and Vetter2013); (6) Dijkman & Kromkamp, (Reference Dijkman and Kromkamp2006); (7) Mansour et al. (Reference Mansour, Frampton, Nichols, Volkman and Blackburn2005); (8) Temperoni (Reference Temperoni2015); (9) this study.
a Drepanopus forcipatus + Calanoides carinatus.
Discussion
Fatty acids have been extensively used to assess trophic preferences in food webs worldwide (reviewed by Dalsgaard et al., Reference Dalsgaard, St. John, Müller-Navarra and Hagen2003), including few studies in the Argentinean Exclusive Economic Zone focused on plankton (Napolitano et al., Reference Napolitano, Pollero, Gayoso, MacDonald and Thompson1997; Dutto et al., Reference Dutto, Kopprio, Hoffmeyer, Alonso, Graeve and Kattner2014). On the contrary, the FATM approach has been scarcely applied on fish early stages (e.g. St. John & Lund, Reference St. John and Lund1996; Rossi et al., Reference Rossi, Sabatés, Latasa and Reyes2006). In this sense, our data set is the first regarding fatty acid profiles of Merluccius hubbsi larvae and their copepod prey in the AEEZ. Data represent valuable trophic signatures in the pelagic food web of the north Patagonian shelf and set a milestone for new analyses regarding not only hake but also other important fish and zooplankton species in the region.
Fatty acid profiles of hake larvae and their copepod prey, dominated by the SFAs 16:0 and 18:0, the MUFAs 18:1n-9 and 22:1n-9, and the PUFA 22:6n-3, were similar to those reported for larvae of Merluccius paradoxus, M. capensis (Grote et al., Reference Grote, Hagen, Lipinski, Verheye, Stenevik and Ekau2011), Gadus morhua and G. macrocephalus (Laurel et al., Reference Laurel, Copeman, Hurst and Parrish2010). Regarding copepods, a large pool of literature on fatty acids composition exists, but it was derived primarily from high latitude species (Falk-Petersen et al., Reference Falk-Petersen, Sargent, Lønne and Timofeev1999). In spite of this, the usually high presence of long-chain monoenes in calanoids was also observed in copepods of the north Patagonian shelf, represented by erucic acid (22:1n-9). The common calanoid marker 20:1n-9 was not detected in high proportions, which could be explained considering that this fatty acid is usually abundant in cold water species that have wax esters as their main lipid storage (Sargent & Falk-Petersen, Reference Sargent and Falk-Petersen1988). Overall, our copepods profiles fairly match results previously obtained for genera Drepanopus (Cripps & Atkinson, Reference Cripps and Atkinson2000) and Calanoides (Falk-Petersen et al., Reference Falk-Petersen, Sargent, Lønne and Timofeev1999). It is worth noting the low proportions of EPA (20:5n-3) observed in larvae and copepods, which is usually a dominant PUFA. It has been suggested that this FA would be physiologically less important than DHA (Watanabe et al., Reference Watanabe, Izquierdo, Takeuchi, Satoh and Kitajima1989). Low values could also derive from the utilization of this FA as an energy source or from its elongation and posterior desaturation to DHA (Veloza, Reference Veloza2005), considering the higher requirement of the latter (Watanabe, Reference Watanabe1993).
Hake larval FA profiles reasonably resembled those of their copepod prey, reinforcing the potential of the fatty acids approach to demonstrate feeding preferences (St. John & Lund, Reference St. John and Lund1996). In agreement, comparison of hake larvae profiles with those from other potential prey from the literature, such as diatoms, dinoflagellates or ciliates evidenced a clear similarity with copepods over other options. However, this remains to be tested with field samples from the north Patagonian shelf. It is worth noting the dominance of DHA both in larvae and copepods, which suggests an accumulation of this FA through dietary intake from copepod prey. Although some alterations in FA can occur from one trophic level to the next, valuable dietary information can be retained despite these metabolic modifications (Dalsgaard et al., Reference Dalsgaard, St. John, Müller-Navarra and Hagen2003). The high percentages of DHA and low proportions of its precursors such as linoleic (18:2n-6) and linolenic (18:3n-3) acids in hake larvae profiles also support the fact that a scarce or null de novo PUFA synthesis might be occurring, with a concurrent lack of modification of dietary PUFA after consumption. Such precursors generally occur in percentages <2% in fish (Ackman, Reference Ackman and Connell1980). On the other hand, the trend in the study area towards the accumulation of DHA from low to high trophic levels (copepods to hake larvae), as discussed by Ackman (Reference Ackman2004) and observed in other fish species (e.g. Rossi et al., Reference Rossi, Sabatés, Latasa and Reyes2006), might be critical for energy storage and cell-tissue development during hake early development, turning into an interesting fitness indicator.
Detection of FA markers is enhanced if higher trophic levels feed extensively on the foods investigated, and samples are taken during a period of anabolism rather than catabolism (St. John & Lund, Reference St. John and Lund1996; Dalsgaard & St. John, Reference Dalsgaard and St. John2004). In this sense, hake larvae are known to be specialist predators upon calanoid copepods (Temperoni & Viñas, Reference Temperoni and Viñas2013), and within the size range included in this study, individuals have high growth rates involved in synthesizing new body structures (Betti et al., Reference Betti, Machinandiarena and Ehrlich2009). Also, environmental variability can alter physiological responses of organisms masking trophic links. However, fatty acid compositions have been reported to be stable unless environmental conditions changed noticeably (Rossi et al., Reference Rossi, Sabatés, Latasa and Reyes2006). Since sampling occurred within a narrow temporal window (January), where such conditions are supposed to remain nearly stable, FA composition was not expected to undergo changes that would mask trophic markers.
With this in mind, particular FA markers were identified in hake larvae and their copepod prey in the NPS. First, the ratio 18:1n-9/18:1n-7 > 1 confirmed carnivorous feeding in hake larvae. Carnivory occurred mainly upon copepods, due to high proportions of 22:1n-9. Odd-chain FA such as 15:0 and 17:0, characteristic of bacteria, were also observed in larvae and copepods profiles. These markers suggest a microbial input at the base of the NPS food web in summer, in agreement with previous analyses based on stable isotopes (Gaitán, Reference Gaitán2012). How can these bacterial markers reach larval tissues? We postulate that protozoans such as heterotrophic dinoflagellates (identified from the 18:4n-3 and 22:6n-3 markers) could be an intermediate step between bacteria and upper trophic levels. Prior studies indicate their prevalence in the nursery area in summer over diatoms (Carreto et al., Reference Carreto, Carignan, Montoya, Cucchi Colleoni, Sánchez and Bezzi2007 and references), which also explains why diatom markers (such as a 16:1/16:0 > 1 ratio) were not identified in high proportions in the profiles. Since dominant herbivorous copepods D. forcipatus and C. carinatus in the NPS (Temperoni et al., Reference Temperoni, Viñas, Martos and Marrari2014) can prey upon dinoflagellates (Antacli, Reference Antacli2011; Santos B., personal communication), they would be transferring bacterial and phytoplankton markers to hake larvae, hence connecting microbial and herbivorous components of the local food web. Due to their intermediate size, the mechanism of trophic upgrading by these protozoans may bridge the gap of essential nutrients (i.e. minerals, vitamins, amino acids, fatty acids and sterols) between the microbial loop and higher trophic levels, as suggested by Klein Breteler et al. (Reference Klein Breteler, Schogt, Baas, Schouten and Kraay1999). Another possible way might be that hake larvae prey upon protozoans directly, considering the high contribution of the dinoflagellate marker DHA to their fatty acids profiles with respect to copepods, and the highest effect of this fatty acid on the MDS sample ordination. There is growing evidence that these organisms, which are generally undetectable with standard gut content studies (particularly naked ones), can play an important role in fish larvae nutrition (Fukami et al., Reference Fukami, Watanabe, Fujita, Yamaoka and Nishijima1999; Overton et al., Reference Overton, Meyer, Støttrup and Peck2010). In this sense, the fatty acids approach could broaden the known trophic spectrum of hake larvae. Even though hake larvae profiles were not very similar to those from dinoflagellates taken from literature, this hypothesis remains to be confirmed with field plankton samples collected in the north Patagonian shelf.
Acknowledgements
We thank the crew and scientific staff on board the RV ‘Eduardo Holmberg’ for their assistance and sample collection during cruises. We are indebted to Gustavo Macchi, head of the M. hubbsi Patagonian Stock Recruitment Project of INIDEP. Constructive input from anonymous reviewers is greatly appreciated.
Financial support
This study was partially supported by funds from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) PIP 112-20110100892 and doctoral fellowships granted to BT, Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) PICT 2013-1484, and Universidad Nacional de Mar del Plata (UNMdP) Projects 15/E667 and 15/E572. This is Instituto Nacional de Investigación y Desarrollo Pesquero (INIDEP) contribution no 2136.