Introduction
Ascarid parasites infect a variety of hosts, including humans (Ascaris lumbricoides Jourdan et al., Reference Jourdan, Lamberton, Fenwick and Addiss2018), swine (A. suum, Thamsborg et al., Reference Thamsborg, Nejsum, Mejer and Holland2013), companion animals (Toxocara spp., Overgaauw, Reference Overgaauw1997), poultry (Ascaridia galli, Kilpinen et al., Reference Kilpinen, Roepstorff, Permit, Norgaard-Nielsen, Lawson and Simonsen2005) and horses (Parascaris spp., Nielsen, Reference Nielsen2016), where Toxocara spp. and A. suum can result in zoonotic transmissions. Infections are commonly associated with wasting disease in children (Jourdan et al., Reference Jourdan, Lamberton, Fenwick and Addiss2018), decreased productivity in livestock (Kilpinen et al., Reference Kilpinen, Roepstorff, Permit, Norgaard-Nielsen, Lawson and Simonsen2005; Thamsborg et al., Reference Thamsborg, Nejsum, Mejer and Holland2013) and stunted growth and intestinal obstruction in companion animals (Overgaauw, Reference Overgaauw1997; Nielsen, Reference Nielsen2016). This array of clinical diseases warrants the routine use of anthelmintics for therapeutic and preventative measures.
Anthelmintic resistance is prevalent and of grave concern among strongylid parasite species (Kaplan, Reference Kaplan2004), but the Ascarididae family has received less attention on this issue. Parascaris spp. is the only example of an ascarid parasite with major issues of anthelmintic resistance (Lyons et al., Reference Lyons, Tolliver, Ionita and Collins2008, Reference Lyons, Tolliver, Kuzmina and Collins2011; Armstrong et al., Reference Armstrong, Woodgate, Gough, Heller, Sangster and Hughes2014; Peregrine et al., Reference Peregrine, Molento, Kaplan and Nielsen2014; Martin et al., Reference Martin, Höglund, Bergström, Karlsson Lindsjö and Tydén2018), as only single case reports of failed anthelmintic efficacy exist for other ascarid species (Yazwinski et al., Reference Yazwinski, Tucker, Wray, Jones and Clark2013; Krücken et al., Reference Krücken, Fraundorfer, Mugisha, Ramünke, Sifft, Geus, Habarugira, Ndoli, Sendegeya, Mukampunga, Bayingana, Aebischer, Demeler, Gahutu, Mockenhaupt and von Samson-Himmelstjerna2017). This unique but troubling status of Parascaris spp. provides an opportunity for identifying drug resistance and metabolism mechanisms that could be extrapolated to other ascarid parasites before resistance levels rise to become a major problem in those as well. Unfortunately, little work has been done to unravel drug response mechanisms for Parascaris spp. Currently, only one published report exists (Janssen et al., Reference Janssen, Krücken, Demeler, Basiaga, Kornaś and von Samson-Himmelstjerna2013), but work is currently being carried out by other investigators and some of this was recently presented at the 2019 World Association for the Advancement of Veterinary Parasitology (W.A.A.V.P.) conference (personal communications, Alexander Gerhard, Nicolas Lammassiaude, Frida Martin and Eva Tydén). The apparent slow progress is due to numerous challenges when studying Parascaris spp. Physical challenges include obtainment of parasites as they are only intermittently established in foals, variation in worm viability/lifespan when harvested from different equine hosts as the host immune system effects on worm viability are unknown, a lack of in vitro protocols for parasite maintenance (nutritional and environmental requirements), difficulties maintaining adult parasites in vitro due to their large size, a lack of in vitro anthelmintic exposure protocols (drug concentration and duration of exposure) and understanding how this drug exposure differs from in vivo drug exposure. Molecular challenges include the absence of a fully annotated Parascaris spp. genome/transcriptome, lack of understanding how the transcriptome differs between parasitic stages/tissues and sex and which of these are most relevant to drug metabolism and resistance, distant phylogenetic relationships between Parascaris spp. and more commonly studied nematodes (Haemonchus contortus, Caenorhabditis elegans), inadequate knowledge of genes (HKGs) and how they might vary between parasite stages and drug exposures, how the drugs enter the parasite (orally or transcuticularly), and the mode of action of anthelmintic drugs and involvement of drug metabolizing enzymes. While the challenges seem endless, recent and current studies are making progress by establishing in vitro maintenance protocols for adult (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018) and larval stages (personal communication, Eva Tydén), availability of a partially annotated genome (Wang et al., Reference Wang, Gao, Mostovoy, Kang, Zagoskin, Sun, Zhang, White, Easton, Nutman, Kwok, Hu, Nielsen and Davis2017), identification of p-glycoproteins in various tissues (Chelladurai and Brewer, Reference Chelladurai and Brewer2019) and preliminary evidence of genes involved with drug resistance and drug exposure (Janssen et al., Reference Janssen, Krücken, Demeler, Basiaga, Kornaś and von Samson-Himmelstjerna2013; personal communications Alexander Gerhard, Nicolas Lamassiaude and Frida Martin).
Leading researchers have suggested that studying drug responses at the transcriptomic level in known susceptible isolates will enhance the understanding of anthelmintic resistance (Beech et al., Reference Beech, Skuce, Bartley, Martin, Prichard and Gilleard2011; Kotze et al., Reference Kotze, Hunt, Skuce, von Samson-Himmelstjerna, Martin, Sager, Krücken, Hodgkinson, Lespine, Jex, Gilleard, Beech, Wolstenholme, Demeler, Robertson, Charvet, Neveu, Kaminsky, Rufener, Alberich, Menez and Prichard2014). Utilizing susceptible isolates may reduce challenges due to genetic variation as resistant isolates have increased genetic diversity (Beech et al., Reference Beech, Skuce, Bartley, Martin, Prichard and Gilleard2011; Kotze et al., Reference Kotze, Hunt, Skuce, von Samson-Himmelstjerna, Martin, Sager, Krücken, Hodgkinson, Lespine, Jex, Gilleard, Beech, Wolstenholme, Demeler, Robertson, Charvet, Neveu, Kaminsky, Rufener, Alberich, Menez and Prichard2014). Xenobiotic defence mechanisms and/or drug responses of parasites are not widely known, and it is uncertain if these defence mechanisms are naturally occurring or developed as a result of drug selection and anthelmintic resistance. Current researchers are exploring the transcriptomic response of parasites belonging to the order Ascarididae following in vitro drug exposure; however, to the authors' knowledge, no results have been published. The unique albeit widespread macrocyclic lactone (ML) resistance status of Parascaris spp. urges the need for uncovering this parasite's response to xenobiotics, which may provide valuable insight to preserve anthelmintic efficacy for other ascarid species and beyond. While several investigators are exploring the effects of in vitro anthelmintic exposure on Parascaris spp. (personal communications Alexander Gerhard, Nicolas Lamassiaude and Frida Martin), there is only one published report (Janssen et al., Reference Janssen, Krücken, Demeler, Basiaga, Kornaś and von Samson-Himmelstjerna2013), but the phenotypic and transcriptomic responses to various drug concentrations are unknown. Only recently has an in vitro maintenance and viability assessment method been tested and established for intestinal stages of Parascaris spp. (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018), and this provides a basis for establishing an in vitro anthelmintic exposure method while monitoring the phenotypic and transcriptomic responses.
The purpose of this study was to aid in overcoming some of the previously mentioned challenges by (1) establishing a method for in vitro anthelmintic exposure of adult Parascaris spp. using different concentrations of anthelmintic drugs, (2) reporting the phenotypic responses to anthelmintic exposure in terms of worm viability over time using a previously established objective scoring system (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018), and (3) providing a preliminary examination of transcriptomic responses of drug-susceptible adult Parascaris spp. to ivermectin (IVM), an ML drug to which resistance is widely established on managed horse farms, and to oxibendazole (OBZ), a drug that remains effective.
Materials and methods
Study design
This study consisted of two parts. Part 1 was to perform in vitro drug exposures at varying concentrations and observe the worm responses using a motility-based scoring system (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018). Part 1 ended with the determination of the sub-lethal concentration for each drug type and exposure length where worm viability decreased approximately ⩾25%. In Part 2, these pre-determined parameters were used for a second in vitro drug exposure. Subsequently, RNA sequencing and gene expression analysis was used to provide a preliminary report of changes in gene expression following drug exposure compared to both in vitro and in situ controls. An illustration of the study design can be found in Fig. 1.

Fig. 1. An illustration of the study design (IVM, ivermectin; OBZ, oxibendazole).
Parasite sources
The study took place over the course of four foal necropsies from December 2017 to September 2018. The foals were born in a herd housed at the University of Kentucky that has not been treated with any anthelmintics since 1979 and have been documented harbouring a variety of equine parasites (Lyons et al., Reference Lyons, Drudge and Tolliver1990). Foals were humanely euthanized following the American Veterinary Medical Association guidelines for the euthanasia of animals when they reached 4.5–5 months old and subsequently necropsied. The research was conducted under the approval from the University of Kentucky's Institutional Animal Care and Use Committee under protocol number 2012-1046.
Collection of Parascaris spp.
Collection of live worm specimens at necropsy occurred as previously described (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018). Brief details are provided in the Supplementary files. For Part 2 of this study (see Fig. 1), additional worm specimens for in situ controls were obtained by leaving numerous worms within a 30 cm section of the jejunum. Intestinal content was allowed to remain in the segment and both ends were tied shut with string. The segment was placed in a closed container and then into the water bath to maintain its temperature at 37 °C. The purpose of the in situ controls was to mimic the natural environment of the worms while minimizing disturbances. Therefore, this in situ control was used as a comparison for the in vitro non-drug-treated controls.
In vitro maintenance and viability assessment of Parascaris spp.
Worms for in vitro drug exposure were maintained in RPMI 1640 medium (R8758, Sigma-Aldrich, St. Louis, MO, US) within TPP tissue culture flasks (300 cm2, MidSci, St. Louis, MO) at 37 °C as described by Scare et al. (Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018). Media were changed every 12 h (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018). Worm viability was assessed at regular time intervals using a motility-based objective scoring system on a 0–6 scale as previously described (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018).
Anthelmintics
The anthelmintics employed in this study were powder formulations of IVM (22,23-dihydroavermectin B1, Sigma-Aldrich) and OBZ (methyl carbamate, Sigma-Aldrich). Stock solutions of both drugs were individually prepared as described by Hu et al. (Reference Hu, Ellis, Yiu, Miller, Urban, Shi and Aroian2013), where 11.4 μ m IVM and 40.1 μ m OBZ (both 100 μg mL−1) were individually dissolved in 100% dimethyl sulfoxide (DMSO). Tenfold serial dilutions were carried out using 10% DMSO to achieve the concentrations of 0.1, 1.0, and 10.0 and 100.0 μg mL−1. These concentrations were based on previous studies using similar concentrations (Hanser et al., Reference Hanser, Mehlhorn, Hoeben and Vlaminck2003; Hu et al., Reference Hu, Ellis, Yiu, Miller, Urban, Shi and Aroian2013; Janssen et al., Reference Janssen, Krücken, Demeler, Basiaga, Kornaś and von Samson-Himmelstjerna2013).
Four millilitres of the prepared drug suspensions were added to the pre-assigned flasks containing 196 mL of RPMI-1640 media so that the final concentration of DMSO in the flask was 0.2% and the drug suspension was 1/50 of the final volume (Hu et al., Reference Hu, Ellis, Yiu, Miller, Urban, Shi and Aroian2013). Control flasks containing only 0.2% DMSO were also prepared. Worms were allowed a 24 h acclimation period before the anthelmintics were added. Drug treatments were applied at every medium change thereafter.
Part 1: initial assessment of parasite responses to in vitro drug exposure
Part 1 was dedicated to observing worm viability in response to in vitro drug exposure at varying concentrations of IVM and OBZ anthelmintics to determine optimal sub-lethal drug concentrations and length of exposure. The number of worms evaluated for each drug concentration can be found in Table 1. Viability assessments as described by Scare et al. (Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018) occurred at the following time points: 0, 1, 2, 3, 4, 6, 12, 18, 24, 30, 42, 54, 66 and 78 h.
Table 1. Number of Parascaris spp. worms used to observe responses to drug exposure in vitro for Part 1 of this study

IVM, ivermectin; OBZ, oxibendazole; Imm, immature worms; RPMI-1640, Roswell Park Memorial Institute-1640 medium; DMSO, dimethyl sulfoxide.
Drugs were prepared in 10% DMSO. Worms were maintained in 200 mL RPMI-1640 media at 37 °C.
The drug concentrations and time points for future snap freezing and transcriptomic analysis (Part 2) were determined when mean worm viability decreased by approximately ⩾25%, but remained sub-lethal. The final determined duration of exposure was extended by 2 h for IVM and 12 h for OBZ to ensure that the decrease in viability was stable, and not due to variation between specimens.
Part 2: RNA-sequencing analysis
In Part 2, a second drug trial was performed using worms harvested from a second necropsy. Subsequently, these specimens were used for RNA-seq analysis. Only adult worms were used due to the lack of immature worms present. Worms were maintained in vitro in groups of four, consisting of two adult males and two adult females. The drug exposure parameters determined from Part 1 were applied. One male and one female for each drug treated/control group were used for further analysis. At the predetermined time points, these worms were snap frozen live in liquid nitrogen and kept at −80 °C until use.
RNA isolation, library preparation and RNA sequencing
Frozen whole worms were ground into a fine powder using a mortar and pestle while continuously adding small amounts of liquid nitrogen. Approximately 100 mg of worm powder was used for RNA isolation which was carried out using TRIzol RNA isolation reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Next, DNase treatment was performed (DNA-free DNA removal kit, Thermo Fisher Scientific). RNA quantity and quality was determined at the University of Kentucky Genomics Core Lab using the Agilent bioanalyser (Agilent, Santa Clara, CA, USA).
RNA samples were sent to the University of Louisville CGeMM DNA Core Facility (http://louisville.edu/genetics/gemm-dna-facility-core) for library preparation and sequencing. Libraries were prepared with Illumina's TruSeq stranded total RNA library kit (Illumina, San Diego, CA, USA) with Ribo-Zero Gold depletion. Libraries were sequenced using the NextSeq 500 High Output v2 75 cycles kit. Samples were run in 1 × 75 base pair configuration, generating up to 400 million reads total, approximately 40 million reads per sample.
RNA-seq analysis and selection of genes of interest
Specific details pertaining to RNA-seq analysis can be found in the Supplementary files. Reads were aligned and annotated to the Parascaris univalens reference genome and transcriptome, respectively (Wang et al., Reference Wang, Gao, Mostovoy, Kang, Zagoskin, Sun, Zhang, White, Easton, Nutman, Kwok, Hu, Nielsen and Davis2017). Mapping statistics are shown in Supplementary Table S1. The RNA sequencing data from this study were deposited in the Gene Expression Omnibus (GEO, NCBI, NIH) database under study GSE129514.
Functional annotation of the differentially expressed genes based on gene ontology (GO; biological process and molecular function) was performed using Pantherdb.org, a pathway analysis program (Mi et al., Reference Mi, Muruganujan, Casagrande and Thomas2013, Reference Mi, Huang, Muruganujan, Tang, Mills, Kang and Thomas2017).
Statistical analyses
Part 1: initial assessment of parasite responses to in vitro drug exposure
Mean viability per flask of worms at each timepoint was calculated using the following formula (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018):

Mean scores and 95% confidence intervals were determined using Microsoft Excel 2016 (Redmond, WA, USA). Further statistical analysis was carried out using SAS software (version 9.4, SAS Institute, Cary, North Carolina, USA). Two mixed-model analyses with repeated measures over time were performed to examine the effects of worm stage (immature/adult) and sex (male/female), and the effects of each drug (IVM or OBZ) at different concentrations on worm viability. The details for the covariates examined and random effects can be found in the Supplementary file materials and methods section.
For both analyses described above, covariates identified as significant (α = 0.05) were further examined in a ‘least squares means’ analysis for a Tukey's pairwise comparison.
Part 2: RNA-sequencing analysis
Identification of differentially expressed genes was performed using a total of five one-way ANOVA analyses, using the Benjamini–Hochberg correction for false discovery rate (P < 0.01). Analyses were performed using JMP software (JMP®, Version 13. SAS Institute Inc., Cary, NC, USA, 1989–2019). The first four analyses ignored the influence of worm sexes. The first analysis considered all treated worms, regardless of drug used, vs all control worms, regardless of in situ or in vitro. The second analysis compared only the IVM-treated and IVM-control worms, and the third analysis compared only the OBZ-treated and OBZ-control worms. The fourth analysis compared all in vitro controls to all in situ control worms. Finally, the fifth analysis ignored drug/control group and considered any differences between worm sexes.
Because small sample sizes were a limitation of this study, the comparison of primary interest was between all drug-treated (OBZ and IVM; n = 4) and all control (in vitro and in situ controls; n = 6) worms. The functionality of the top ten significantly different genes within this comparison was explored using Wormbase (Parasite.Wormbase.org) and NCBI. Lastly, due to a lack of verified HKGs for Parascaris spp., two commonly used HKGs, namely nuclear cap-binding protein subunit 2 (ncbp) and RNA polymerase RPABC1 large subunit (ama), were observed for stable expression (Fragments Per Kilobase of transcript per Million mapped reads; FPKM values) between the different drug treatments and corresponding controls, in vitro and in situ controls, and all treated and all controls. These HKGs were selected based on a previous study examining HKG expression in treated H. contortus (Lecová et al., Reference Lecová, Růžičková, Laing, Vogel, Szotáková, Prchal, Lamka, Vokřál, Skálová and Matoušková2015).
Functional annotation of the differentially expressed genes based on GO (biological process and molecular function) was performed using Pantherdb.org, a pathway analysis program (Mi et al., Reference Mi, Muruganujan, Casagrande and Thomas2013, Reference Mi, Huang, Muruganujan, Tang, Mills, Kang and Thomas2017).
Results
Part 1: initial assessment of parasite responses to in vitro drug exposure
A graphical representation of worm viability following in vitro drug exposure can be found in Fig. 2. Overall, IVM had a more immediate effect on worm viability than OBZ.

Fig. 2. A graphical representation of mean worm viability following in vitro anthelmintic exposure. Control worms were maintained in RPMI 1640 medium only or with dimethyl sulfoxide (DMSO) which was used to prepare the anthelmintics. Error bars represent 95% confidence intervals (α = 0.05).
Neither the worm stage nor sex had a significant influence on viability (P = 0.1868). Between the two drug treatments, worms exposed to IVM at 0.1 μ m (0.1 μg mL−1) had significantly lower viability than the RPMI-1640 control worms at hours 3 and 4. No other significant differences were found between groups for the other time points, including the RPMI-1640 (no drug control) and the DMSO 10% control, which indicates that the DMSO did not affect worm viability and any observed changes were due to the drug.
Because of the lack of significant differences in worm viability between the three IVM concentrations tested, 1.1 μ m (1 μg mL−1) was chosen because it was the median concentration employed. Due to the lack of significant differences in worm viability between all OBZ concentrations and control worms, the highest OBZ concentration of 40.1 μ m (10 μg mL−1) was chosen for Part 2. Viability had decreased by ⩾25% for all IVM concentrations at 1 h post-treatment and at 12 h for OBZ at 40.1 μ m (10 μg mL−1). The final timepoints for Part 2 were extended by 2 h for IVM and 12 h for OBZ to ensure a representative decrease in viability rather than inter-specimen variation for final exposure lengths of 3 and 24 h, respectively.
Part 2: RNA-sequencing analysis
Figure 3 illustrates the viability of the worms collected from the second necropsy and subsequently used for RNA sequencing. The total number of genes with significantly different expression levels (α = 0.01) for each of the group comparisons can be found in Table 2. A list of all the significantly different genes for each comparison is available in the Supplementary files. Figure 4 shows the expression of two HKGs, ncbp and ama, and four other genes identified as having potential functional relevance that were significantly different between the treated and control worms. These genes were cytochrome p450 4c1 (cyp4504c1), frmd4a, klhdc10 and sup-9.

Fig. 3. A graphical representation of mean worm viability of worms used in Part 2 of this study, where those exposed to oxibendazole (OBZ) at 40.1 μ m (10 μg mL−1) for 24 h and ivermectin (IVM) at 1.1 μ m (1 μg mL−1) for 3 h were used for RNA-sequencing analysis. Control worms were maintained in RPMI 1640 medium only or with dimethyl sulfoxide (DMSO) which was used to prepare the anthelmintics. Error bars represent 95% confidence intervals (α = 0.05).
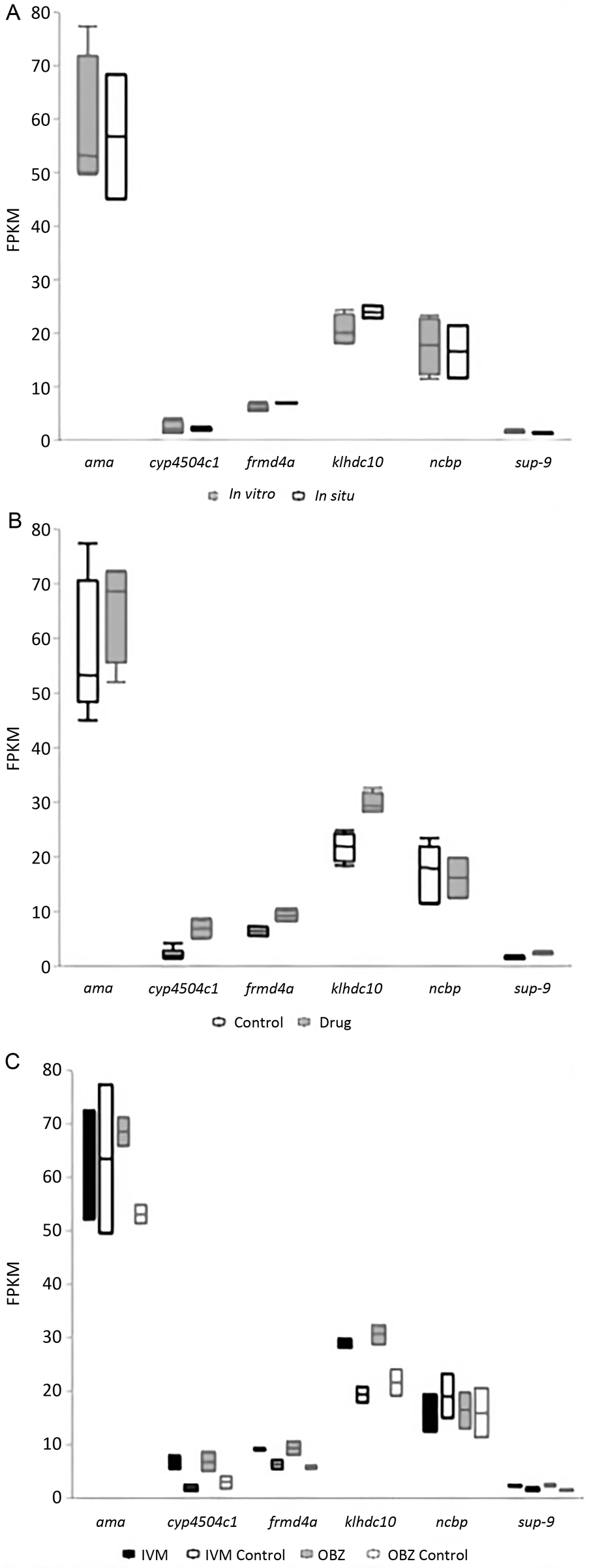
Fig. 4. A graphical representation of select genes from the RNA-seq analysis performed in Part 2 of this study. The housekeeping genes are ama and ncbp. (A) In vitro vs in situ controls, (B) all control worms vs all treated worms, where the selected genes had significantly higher expression in the treated worms (α = 0.01), (C) ivermectin (IVM) treated with 1.1 μ m (1 μg mL−1), IVM control, oxibendazole (OBZ) treated with 40.1 μ m (10 μg mL−1), and OBZ control.
Table 2. The number of significant transcripts (α = 0.01) based on FPKM values obtained from Part 2 of this study

OBZ, oxibendazole; IVM, ivermectin.
a All target genes had significantly higher expression in drug-treated worms than control worms.
For the GO analysis, the entire list of genes from the annotated genome (Wang et al., Reference Wang, Gao, Mostovoy, Kang, Zagoskin, Sun, Zhang, White, Easton, Nutman, Kwok, Hu, Nielsen and Davis2017) was initially used. Of these, 6% did not have a listed gene product and 24% were considered annotated as a ‘hypothetical protein’, and were not included in the GO analysis. The following comparisons were made: all treated vs all controls, IVM 1.1 μ m (1 μg mL−1) vs IVM controls, and OBZ 40.1 μ m (10 μg mL−1) vs OBZ controls. The significantly different genes corresponding to these comparisons are listed in the Supplementary files. Predicted biological processes and molecular function pathways are presented in Fig. 5. Overall, it appears that treatment with IVM reduced the number of categories for genes involved with molecular functions and biological processes. However, the categories of genes following OBZ treatment were similar to the gene categories representing all of the genes of the control worms.

Fig. 5. A graphical representation of the gene ontology pathway analysis for significantly different genes (SDGs) between groups. From left to right: all genes, all treated worms vs all control worms, ivermectin (IVM) treated with 1.1 μ m (1 μg mL−1) vs IVM control, oxibendazole (OBZ) treated with 40.1 μ m (10 μg mL−1) vs OBZ control. The top row reflects biological processes (BP) and the bottom row reflects molecular functions (MF).
Discussion
This study addresses some of the challenges associated with in vitro drug exposure of adult Parascaris spp. First, it has established a method for in vitro anthelmintic exposure of adult Parascaris spp. Second, it is the first report to objectively describe the phenotypic responses (viability) of anthelmintics with different modes of action at varying concentrations. Third, it provides a reference for using in situ controls which may better represent in vivo conditions rather than in vitro controls. Lastly, this report also provides preliminary data for a whole-transcriptome analysis of anthelmintic-naïve Parascaris spp. following in vitro drug exposure.
Prior to this study, there was no comparison of different anthelmintics at varying drug concentrations or exposure lengths for in vitro anthelmintic exposure of any adult ascarid parasite. Only one published report of in vitro drug exposure of Parascaris spp. exists employing IVM at 10−8 or 10−9 M for 12 h (Janssen et al., Reference Janssen, Krücken, Demeler, Basiaga, Kornaś and von Samson-Himmelstjerna2013), but no reference was provided for these parameters and the phenotypic effects were unknown. Therefore, the anthelmintic drug concentrations (0.1, 1.0 and 10 μg mL−1) chosen for the current study were based on a previous study examining in vitro anthelmintic effects of various nematode parasites, including immature Ascaris suum (Hu et al., Reference Hu, Ellis, Yiu, Miller, Urban, Shi and Aroian2013). It is also important to note that the method of drug preparation by dissolving powder formulations in DMSO allowed the drug to affect the worms and the DMSO did not influence the worm viability (Fig. 2). The phenotypic responses, displayed by worm viability, were different between IVM and OBZ (Fig. 2) and correspond to the drugs' mode of action. Overall, the phenotypic effects of the different IVM concentrations on worms from the first necropsy did not differ significantly between stages/sexes, and decreases in viability were observed within 1 h. However, the effects of OBZ were more prominent in the immature worms, and not significantly different from the control worms for any stage/sex. Interestingly, there does not appear to be a dose-dependent effect on viability (Figs 2 and 3). The cause of this is unknown, but may be due to a number of reasons such as a lack of proper nutrition for the worms causing unstable viability, misinterpretations of drug bioavailability in Parascaris spp., the drug concentrations employed may be too high (IVM) or too low (OBZ), or perhaps there is too much interspecimen variation even when worms are harvested from the same foal.
The effects on viability reflect each drugs' mode of action, where IVM is known to have a more immediate paralytic effect (Martin, Reference Martin1997) and OBZ kills worms more slowly by disrupting energy metabolism (Lacey, Reference Lacey1988). Therefore, future studies should examine the effects of OBZ at higher concentrations. Previously, Lloberas et al. (Reference Lloberas, Alvarez, Entrocasso, Ballent, Virkel, Luque, Lanusse and Lifschitz2015) examined IVM concentrations within abomasal digesta and H. contortus following intraruminal IVM administration in lambs. They found IVM present at a concentration of 395 ng g−1 in the abomasal content and at 102 ng g−1 within H. contortus. The drug concentrations employed in the current study are within the same order of magnitude and may be considered physiologically relevant. However, there are some differences that must be considered, such as the different digestive anatomy between ruminants and horses, and the biological differences between H. contortus and Parascaris spp. Therefore, we cannot conclude how the concentrations employed herein would represent in vivo responses. Overall, while the described methodology makes in vitro anthelmintic exposure of adult Parascaris spp. possible, there are still many questions to be answered. Overall, we believe that the drug exposure method described herein will provide a reference for future drug exposure analysis.
One of the primary challenges for in vitro work with Parascaris spp. is the ability to keep worms alive and viable (i.e. retaining motility) for longer than a few days. This challenge has been described by other investigators (personal communications Alexander Gerhard, Nicolas Lamassiaude and Frida Martin) as well as in a previous publication by our group (Scare et al., Reference Scare, Steuer, Shaffer, Slusarewicz, Mousley and Nielsen2018). The quick loss in viability complicates the reliability of worm health in vitro, and comparisons of drug-treated worms to in vitro controls should be interpreted with caution. Instead, it may be more reliable to compare drug treatment to in situ controls. As described above, it is possible to obtain in situ controls by excising a segment of the jejunum (about 30 cm) and tying the ends while worms and intestinal content remain inside. The segment can be incubated at 37 °C. This way, the worms undergo minimal handling/disruption and can remain in a more natural environment during recovery.
Another aim of the study was to provide a preliminary transcriptomic analysis following drug exposure. This was first examined using a GO analysis as described above. The predicted biological functions and metabolic pathways illustrate a variety of processes (Fig. 5) that are presumably affected by anthelmintic exposure. At this time, it is unknown how the differentially expressed genes are influenced by in vitro drug exposure. We also cannot conclude if they would elicit the same response in vivo, or if they have a role in anthelmintic resistance mechanisms. The proportion of significant genes related to various biological processes and molecular functions differ between the IVM-treated/control and OBZ-treated/control comparisons. For the IVM comparison, the genes related to biological processes are only related to metabolic and cellular processes while those related to molecular function are defined as catalytic activity, binding and molecular function regulation (Fig. 5). This is interesting given the IVMs’ paralytic mode of action. The target receptors of IVM (GluCl and GABA channels) are highly expressed on the motor and sensory neurons, which regulate nematode locomotion, feeding behaviour and mediate sensory inputs (Wolstenholme, Reference Wolstenholme2012). The predicted functions appear to be in agreement with the IVM mode of action, such as the disruption of feeding behaviour may be related to changes in metabolic processes, and mediation of sensory inputs likely has some effect on cellular processes, catalytic activity and regulation of molecular function. The BZ mode of action is primarily to disrupt energy metabolism and cell structure (Lacey, Reference Lacey1988). The pathway analysis resulted in a variety of genes, where those related to biological processes were primarily involved with localization, biological regulation, metabolic and developmental processes. The molecular function was dominated by genes related to binding and catalytic activity. The disruption of cellular structure is likely involved in localization, biological regulation, binding and development processes. Likewise, it is not illogical to equate the disruption of energy metabolism with metabolic processes and catalytic activity. Overall, it can be speculated that the predicted GO pathways have some relevance to the drug mode of action, and there are potential pathways involved with drug responses to be explored.
The RNA-sequencing analysis revealed a number of gene transcripts that were significantly different between the treated and control groups (Table 2). Four of the top ten significantly different genes exhibit functions in other organisms related to drug detoxification (cyp4504c1; Feyereisen, Reference Feyereisen1999; Cvilink et al., Reference Cvilink, Lamka and Skálová2009; Yilmaz et al., Reference Yilmaz, Ramünke, Demeler and Krücken2017), coordinating muscle contraction and regulation of membrane potential (Lesage and Lazunski, Reference Lesage and Lazdunski2000; Perez de la Cruz et al., Reference Perez de la Cruz, Levin, Cummins, Anderson and and Horvitz2003), microtubule polymerization (frmd4a; Stehbens et al., Reference Stehbens, Paterson, Crampton, Shewan, Ferguson, Akhmanova, Parton and Yap2006; Ikenouchi and Umeda, Reference Ikenouchi and Umeda2010; Meng and Takeichi, Reference Meng and Takeichi2009) and antioxidant defence mechanisms (klhdc10; James et al., Reference James, Hudson and Davey2009). We hypothesize that these functions may compliment the IVM and OBZ modes of action. Because these genes were not significantly different between the in situ and in vitro controls, it appears that these changes were in fact a response to drug exposure rather than in vitro conditions (Fig. 4A). Although the fold-difference was small (⩽2), all four of these genes were significantly increased among the treated worms (Fig. 4B and C), and it should be noted that even minute changes in RNA expression have been associated with important biological functions (Laurent et al., Reference Laurent, Shtokalo, Tackett, Yang, Vyatkin, Milos, Seilheimer, McCaffrey and Kapranov2013). Given the low sample size (treated worms n = 4; control worms n = 6) and corresponding statistical limitations of Part 2, only speculations rather than conclusions can be made at this time. Interestingly, traditionally researched genes, such as p-glycoproteins (Janssen et al., Reference Janssen, Krücken, Demeler, Basiaga, Kornaś and von Samson-Himmelstjerna2013, Reference Janssen, Krücken, Demeler and von Samson-Himmelstjerna2015; Chelladurai and Brewer, Reference Chelladurai and Brewer2019) and the multi-drug-resistant protein (Kotze et al., Reference Kotze, Hunt, Skuce, von Samson-Himmelstjerna, Martin, Sager, Krücken, Hodgkinson, Lespine, Jex, Gilleard, Beech, Wolstenholme, Demeler, Robertson, Charvet, Neveu, Kaminsky, Rufener, Alberich, Menez and Prichard2014), were not observed to be differentially expressed between drug-treated and control worms.
In summary, this study provides a protocol for in vitro anthelmintic exposure of adult Parascaris spp. and proven useful for phenotypic analyses. It also introduces the concept of using an in situ control as an alternative and potentially more reliable comparison than in vitro controls. While the transcriptomic analysis results are preliminary and should be interpreted with caution, the GO analysis and early transcriptome analysis provide some support that the worms are capable of responding to drug exposure in vitro. This report utilized specimens from an anthelmintic-naïve population which provides a unique perspective as the majority of reports focus on resistant populations. Future studies should investigate the phenotypic and transcriptomic responses of various resistant populations compared to anthelmintic-naïve populations. The work presented herein serves to overcome a few of the known challenges when performing in vitro drug exposure of adult Parascaris spp., but many other existing challenges and the widespread resistance status of Parascaris spp. warrant further investigation of these topics.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182020000189
Acknowledgements
We would like to acknowledge the hard work and dedication by the farm staff in providing the upmost care for this herd of research horses, ‘The Gene Lyons Historic Parasitology Research Herd’, and the supporters of this crowdfunding initiative. We are also grateful for the technical expertise and guidance provided by Dr Richard E. Davis and Dr Jianbin Wang.
Financial support
This research was funded by the Gluck Equine Research Foundation and the Burroughs Wellcome Fund Collaborative Research Travel Award.
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
The research was conducted under the approval from the University of Kentucky's Institutional Animal Care and Use Committee under protocol number 2012-1046.










