Introduction
Weedy ryegrasses (Lolium spp.), including Italian ryegrass [Lolium perenne L. ssp. multiflorum (Lam.) Husnot], rigid ryegrass (Lolium rigidum Gaudin), and perennial ryegrass (Lolium perenne L.), are problematic weeds causing economic losses in U.S. cropping systems, particularly in the Pacific Northwest (Avila-Garcia and Mallory-Smith Reference Avila-Garcia and Mallory-Smith2011; Hulting et al. Reference Hulting, Dauer, Hinds-Cook, Curtis, Koepke-Hill and Mallory-Smith2012; Jasieniuk et al. Reference Jasieniuk, Ahmad, Sherwood, Firestone, Perez-Jones, Lanini, Mallory-Smith and Stednick2008; Nandula et al. Reference Nandula, Reddy, Poston, Rimando and Duke2008; Simarmata et al. Reference Simarmata, Kaufmann and Penner2003). Treatment with PRE and early POST herbicides has been the main means of ryegrass control over the last three decades. As a result, however, resistance to herbicides with different mechanisms of action have evolved and been reported in many ryegrass populations worldwide (Heap Reference Heap2018).
Among the Lolium species, L. perenne ssp. multiflorum is the most economically significant weedy species in annual and perennial cropping systems of California, although L. rigidum has also been documented in the state (DiTomaso and Healy Reference DiTomaso and Healy2007). Herbicide-resistant L. rigidum was first identified in 1998 (Simarmata et al. Reference Simarmata, Kaufmann and Penner2003), where glyphosate, at the labeled field rate, failed to control a population from a sweet almond [Prunus dulcis (Mill.) D. A. Webb] orchard in the northern Sacramento Valley. Glyphosate resistance was later confirmed in several L. perenne ssp. multiflorum populations from a diversity of crops across the Central Valley in 2005 (Jasieniuk et al. Reference Jasieniuk, Ahmad, Sherwood, Firestone, Perez-Jones, Lanini, Mallory-Smith and Stednick2008). Recently, L. perenne ssp. multiflorum populations have evolved resistance to multiple herbicides differing in mechanisms of action (Karn et al. Reference Karn, Beffa and Jasieniuk2018; Tehranchian et al. Reference Tehranchian, Nandula, Jugulam, Putta and Jasieniuk2018). In a previous study, we confirmed multiple resistance to glyphosate, sethoxydim (acetyl CoA carboxylase [ACCase] inhibitor), and paraquat in two L. perenne ssp. multiflorum populations, MR1 and MR2 from an orchard and alfalfa (Medicago sativa L.) field, respectively, in northern California (Tehranchian et al. Reference Tehranchian, Nandula, Jugulam, Putta and Jasieniuk2018). However, resistance to paraquat in MR2 plants was negligible compared with MR1. These populations were also cross-resistant to other ACCase-inhibiting herbicides, including clethodim (only MR1), fluazifop, fenoxaprop, and cyhalofop.
Preliminary greenhouse screening to identify alternative herbicide options to control the MR1 and MR2 populations indicated that both populations were resistant to imazamox and mesosulfuron-methyl (henceforth, referred to as mesosulfuron), both acetolactate synthase (ALS)-inhibiting herbicides, at the labeled field rates. Resistance to ALS inhibitors in Lolium spp. had previously only been reported for a few L. perenne accessions from California roadsides (Saari et al. Reference Saari, Cotterman, Smith and Primiani1992) due to ALS inhibitors not being commonly used in California agriculture. However, recently, ALS inhibitors from the sulfonylurea, imidazolinone, and triazolopyrimidine-sulfonamide families have become increasingly prevalent in tree and vine crops of northern California (Hanson et al. Reference Hanson, Wright, Sosnoskie, Fischer, Jasieniuk, Roncoroni, Hembree, Orloff, Shrestha and Al-Khatib2014).
ALS is the critical enzyme (EC 2.2.1.6) for the biosynthesis of the branched-chain amino acids leucine, valine, and isoleucine in plant species and microorganisms (Whitcomb Reference Whitcomb1999), and is the target site of ALS-inhibiting herbicides. Since the early 1980s, ALS-inhibiting herbicides have been widely used for weed management in agriculture worldwide (Tranel and Wright Reference Tranel and Wright2002; Yu and Powles Reference Yu and Powles2014). Overreliance on ALS-inhibiting herbicides in cropping systems in other regions of the United States and the world has led to the evolution of ALS-inhibitor resistance in more than 159 weed species worldwide (Heap Reference Heap2018). According to Yu and Powles (Reference Yu and Powles2014), the mechanism of ALS-inhibitor resistance in weed populations is either an altered target site and/or a non–target site based mechanism, such as enhanced herbicide metabolism by cytochrome P450 monooxygenases (CYP) or reduced absorption and translocation. To date, an altered target site has been the most common mechanism conferring resistance to the ALS-inhibiting herbicides in weed species (Tranel et al. Reference Tranel, Wright and Heap2018; Yu and Powles, Reference Yu and Powles2014). Several different ALS gene point mutations (Ala-122 to Thr, Tyr, or Val; Pro-197 to Ala, Arg, Asn, Gln, His, Ile, Leu, Ser, or Thr; Ala-205 to Val; Asp-376 to Glu; Arg-377 to His; Trp-574 to Leu; Ser-653 to Thr, Asn, or Ile; and Gly-654 to Asp) have been identified that rapidly cause a moderate to high level of resistance to ALS inhibitors at both the individual and population levels (Tranel et al. Reference Tranel, Wright and Heap2018). Depending on the position of these mutations within the five conserved domains of the ALS gene, different cross-resistance patterns may be endowed in weed species. Despite an altered target enzyme being commonly identified as the mechanism of resistance to the ALS-inhibiting herbicides, other studies have also identified non–target site mechanisms in weed species such as L. rigidum (Busi et al. Reference Busi, Vila-Aiub and Powles2011) and barnyardgrass [Echinochloa crus-galli (L.) P. Beauv.] (Riar et al. Reference Riar, Norsworthy, Srivastava, Nandula, Bond and Scott2013). There are several studies revealing that organophosphate insecticides such as malathion and piperonyl butoxide (PBO) can inhibit herbicide detoxification by CYP (Kreuz and Fonné-Pfister Reference Kreuz and Fonné-Pfister1992). Additionally, it seems that auxinic herbicides such as 2,4-D induce expression of CYP in herbicide-susceptible L. rigidum plants, protecting them against ALS and ACCase inhibitors (Han et al. Reference Han, Yu, Cawthray and Powles2013).
The objectives of this research were to: (1) determine the resistance level to imazamox and mesosulfuron, (2) evaluate other herbicide options available, and (3) investigate the mechanism(s) underlying ALS-inhibitor resistance in multiple herbicide–resistant L. perenne ssp. multiflorum from California.
Materials and methods
Plant material and greenhouse conditions
Two multiple resistant L. perenne ssp. multiflorum populations (MR1, MR2) previously determined to be resistant to glyphosate, ACCase-inhibiting herbicides, and paraquat (Tehranchian et al. Reference Tehranchian, Nandula, Jugulam, Putta and Jasieniuk2018) and a known herbicide-susceptible population (Sus) were investigated for resistance to ALS-inhibiting herbicides in this study. MR1 and MR2 plants that survived 16X (8,960 g ai ha−1) of sethoxydim (hereafter referred to as subpopulations MR1-A and MR2, respectively) and MR1 plants that survived 2X (1,120 g ai ha−1) of paraquat (hereafter referred to as subpopulation MR1-P) in the dose–response experiments conducted for the earlier study (Tehranchian et al. Reference Tehranchian, Nandula, Jugulam, Putta and Jasieniuk2018) were grown in three separate greenhouses at the University of California, Davis, CA. Each greenhouse was set at 25/19 C (day/night) temperatures under a 14-h photoperiod provided by high-pressure sodium lights (400 µmol m−2 s−1). Plants from each subpopulation were allowed to cross-pollinate during March 2018. Seeds were collected at maturity, pooled across plants for each subpopulation (MR1-A, MR1-P, MR2), and stored at 4 C for 2 mo to break dormancy. Based on preliminary greenhouse studies, progeny of these MR subpopulations were resistant to imazamox and mesosulfuron at the labeled field rates when compared with Sus plants.
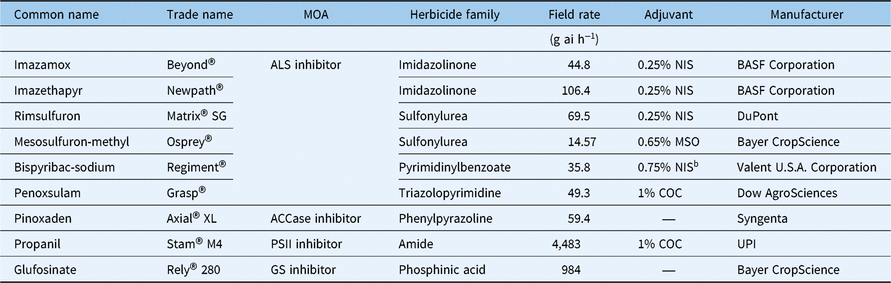
For the following studies, L. perenne ssp. multiflorum seeds from each subpopulation were germinated separately on moist filter paper in petri plates with 1% v/v Captan 80 WDG (Agri Star, Ankeny, IA) and incubated at ambient temperature under a 12-h photoperiod provided by fluorescent lights (160 µmol m−2 s−1). Seedlings at the 1-leaf stage were transplanted into plastic pots (5-cm height by 4.5-cm diameter) filled with commercial potting mix (LC1, Sun Gro Horticulture, Seba Beach, AB, Canada) and maintained in a greenhouse under the conditions described earlier. All herbicide treatments (Table 1) were applied to plants at the 3- to 4-leaf stage (8- to 10-cm height) using an automated track sprayer equipped with an 8001E flat-fan nozzle (TeeJet® Technologies, Springfield, IL) calibrated to deliver 187 L ha−1 at 296 kPa.
Table 1. Herbicides and the labeled field rates used in this studya
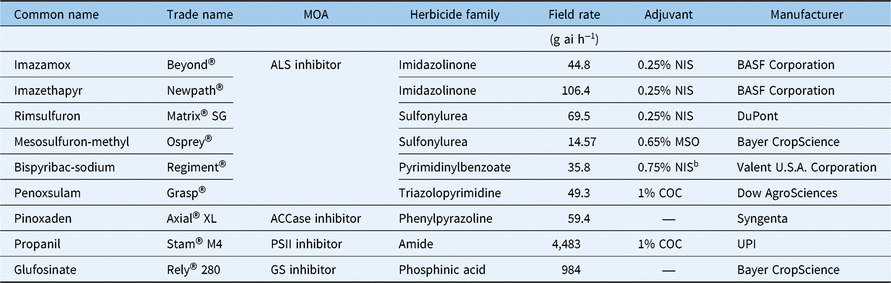
a Abbreviations: ACCase, acetyl CoA carboxylase; ALS, acetolactate synthase; COC, crop oil concentrate; GS, glutamine synthetase; MOA, mechanism of action; MSO, methylated seed oil; NIS, nonionic surfactant; PSII, photosystem II.
b Dyne-A-Pak, nonionic spray adjuvant and deposition aid.
Dose–response experiments
Experiments were conducted in a randomized complete block design (RCBD) with the three MR subpopulations (MR1-A, MR1-P, and MR2) and one Sus population and 25 replications of individuals per ALS-inhibiting herbicide rate. All plants were treated with 0, 1/8, 1/4, 1/2, 1, 2, 4, 8, or 16 times the labeled field rate of imazamox containing 0.25% v/v nonionic surfactant (Induce®, Helena Agri-Enterprises, Collierville, TN ) and mesosulfuron containing 0.65% methylated seed oil (MSO; Southern Ag, Hendersonville, NC), equating to 0, 5.6, 11.2, 22.4, 44.8, 89.6, 179.2, 358.4, or 716.8 g ai ha−1 imazamox and 0, 1.82, 3.64, 7.28, 14.57, 29.14, 58.88, 116.5, or 233 g ai ha−1 mesosulfuron, respectively. Sus plants were treated with 0, 1/32, 1/16, 1/8, 1/4, 1/2, 1, 2, or 4 times the labeled field rate of each herbicide. Plants were maintained in the greenhouse following herbicide treatment under the conditions described above, and plant mortality was recorded at 21 d after treatment (DAT). Mortality data were subjected to probit analysis using PROC PROBIT in SAS (v. 9.4, SAS Institute, Cary, NC) to evaluate the herbicide dose required for 50% plant mortality (LD50). To calculate the resistance index (R/S), the LD50 value of each MR subpopulation was divided by the LD50 of the Sus population.
Cross- and multiple resistance
Experiments with an RCBD factorial with four populations (three MR subpopulations and one Sus population) by seven herbicides were conducted to investigate cross-resistance to other ALS-inhibiting herbicides from four different chemical families and to assess efficacy of alternative herbicides currently available in California. Seedlings at the 3- to 4-leaf stage were treated with the labeled field rate of each herbicide as shown in Table 1 with 10 replications (individuals) per treatment. The experiment was repeated once. Aboveground biomass of all plants was harvested at 21 DAT, oven-dried at 60 C for 48 h, and weighed. Data were subjected to ANOVA using the PROC MIXED procedure in SAS. Due to a nonsignificant experiment effect, data from the two experiments were pooled and the treatment means were separated using Fisher’s protected LSD at α = 0.05.
In vitro ALS enzyme assay
ALS enzyme activity of plants at the 3- to 4-leaf stage was assayed in vitro using procedures similar to previous studies (Nandula and Messersmith Reference Nandula and Messersmith2000; Ray Reference Ray1984). Enzyme was extracted from 4 g of fresh tissue, bulked from 10 to 15 plants, by grinding under liquid nitrogen. The powdered tissue was homogenized in 20 ml of ice-cold buffer (0.1 M potassium phosphate [K2HPO4] at pH 7.5, 1 mM sodium pyruvate, 0.5 mM magnesium chloride [MgCl2], 0.5 mM thiamine pyrophosphate [TPP], 10 µM flavin adenine dinucleotide [FAD], 1 mM dithiothreitol [DTT], 10% v/v glycerol, and 5% w/v polyvinylpolypyrrolidone) for 30 s with a household blender. The homogenate was filtered through eight layers of cheesecloth and centrifuged at 27,000 × g for 15 min. The supernatant was brought to 50% saturation with ammonium sulfate and centrifuged as before. The enzyme pellet was dissolved in 2.5 ml of resuspension buffer (0.1 M K2HPO4, pH 7.5, 20 mM sodium pyruvate, 0.5 mM MgCl2, and 1 mM DTT) and desalted on a Sephadex G-25 column (PD-10 column, Amersham Pharmacia Biotech AB, SE-75 1 84, Uppsala, Sweden) equilibrated with the same buffer. The eluted enzyme was immediately used for assays.
ALS activity was measured by adding 100 µl of enzyme extract to 400 µl of a reaction mix (20 mM K2HPO4, pH 7.0, 20 mM sodium pyruvate, 0.5 mM MgCl2, 0.5 mM TPP, 10 µM FAD, and various concentrations of imazamox or mesosulfuron in a final volume of 0.5 ml) in 1.5-ml tubes. Technical-grade solid imazamox and mesosulfuron were dissolved in an aqueous solution of acetone, and serial dilutions were made to yield final concentrations of 0, 0.0001, 0.001, 0.01, 0.1, and 1 mM in the reaction mix. The reaction was incubated at 37 C for 1 h, then terminated by adding 50 µl of 6 N H2SO4. The acidified reaction mixtures were heated at 60 C for 15 min. This process decarboxylates acetolactate to acetoin. Acetoin was detected as a colored complex after addition of 0.5 ml of 0.5% (w/v) creatine and 0.5 ml of 5% (w/v) α-naphthol (freshly prepared in 2.5 N NaOH) and heating the reaction mixture at 60 C for 15 min (Westerfield Reference Westerfield1945). Absorbance of the colored complex was measured at 525 nm against a no-enzyme blank. ALS activity was expressed as percentage of no herbicide control. The experiment was conducted two times. Each replication indicates one individual extraction. Data were subjected to ANOVA using PROC GLM in SAS. Data from the two runs of the experiment were pooled because there was no significant experimental effect. Nonlinear regression analysis was applied to define a three-parameter power equation of the following form:
to relate the effect of herbicide concentration (x) on ALS activity (y), where y 0 is an asymptote, a is a constant, and b is the slope of the curve. Equation parameters were computed using SigmaPlot (v. 12.5, Systat Software, San Jose, CA).
ALS Sanger sequencing
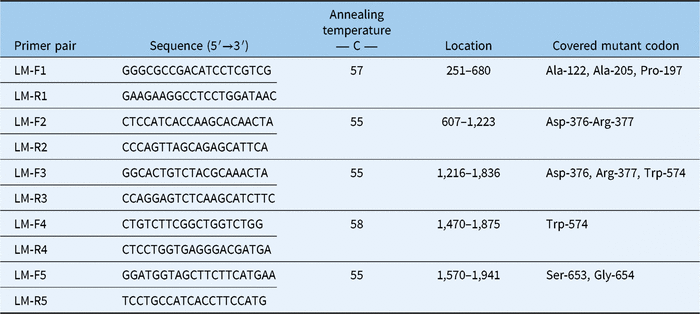
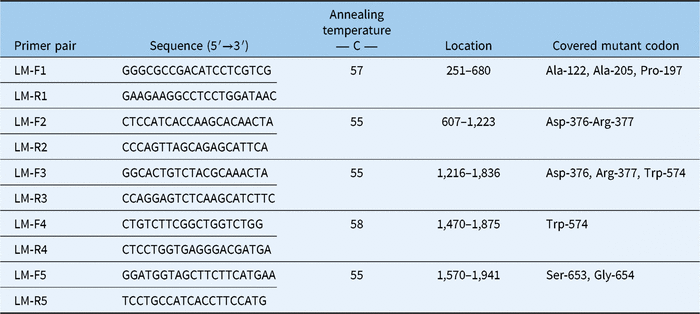
Fresh leaf tissue of four individual plants that survived treatment with both imazamox and mesosulfuron were used for RNA extraction. Additionally, RNA was extracted from leaves of untreated known herbicide-susceptible plants. RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). cDNA was constructed using a Maxima H Minus First Strand cDNA synthesis kit (Thermo Fisher Scientific, Petaluma, CA). To detect mutations in the ALS gene previously identified to confer resistance to ALS-inhibiting herbicides (Yu and Powles Reference Yu and Powles2014), five sets of primers (Table 2) were designed based on the L. multiflorum ALS precursor mRNA complete sequence (NCBI Genbank accession no. AF310684) and L. rigidum biotype 3105–1 ALS gene, partial cds (Genbank accession no. EF411171.1). Primers were synthesized by integrated DNA Technologies (Redwood City, CA). Each polymerase chain reaction (PCR) contained: 12.5 µl of GoTaq Green Master Mix (Promega, Madison, WI), 0.5 µl of each primer (10 µM), ∼16 ng cDNA, and nuclease-free water. The thermal cycler (Bio-Rad T100, Bio-Rad Laboratories, Hercules, CA) was programmed as described in Tehranchian et al. (Reference Tehranchian, Nandula, Jugulam, Putta and Jasieniuk2018) with the primer annealing temperatures shown in Table 2. PCR products were separated on a 1.5% v/v agarose gel and visualized under a UV transilluminator. Amplicons were purified using ExoSAP-ITTM PCR Product Cleanup Reagent (Thermo Fisher Scientific) following the manufacturer’s protocol. The purified PCR products were used in BigDye PCR reactions using the BigDye Terminator Cycle sequencing kit (Thermo Fisher Scientific) and subsequently sequenced with an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). All sequences were aligned and edited using Geneious software (Biomatters, Newark, NJ). Sequences of ALS inhibitor–resistant and ALS inhibitor–susceptible plants were submitted to NCBI Genbank.
Table 2. Primers designed for single-nucleotide polymorphism detection in ALS gene of Lolium perenne ssp. multiflorum (LM)
CYP inhibition by malathion and PBO
No ALS-inhibitor resistance mutations were detected in MR2 plants. Thus, to determine whether resistance to ALS-inhibiting herbicides in MR2 plants is associated with metabolism by CYP, 10 plants at the 3- to 4-leaf stage were treated separately with spray solutions containing 1,000 g ai ha−1 of malathion and PBO. Plant growth and greenhouse conditions were as described earlier. The experiment was conducted in an RCBD with a factorial arrangement of two populations (MR2 subpopulation and Sus), three herbicidal solutions (mesosulfuron, mesosulfuron + malathion, mesosulfuron + PBO) and three malathion/PBO pretreatments (0, 2, 24 h) before herbicide application. The herbicidal solutions were applied at the labeled field rate containing 0.65% MSO. Treatments containing malathion alone, PBO alone, and a nontreated control were also included. To determine the antagonistic effect of 2,4-D on the efficacy of these ALS inhibitors, a separate experiment was conducted. MR2 and Sus seedlings were pretreated with 1,065 g ae ha−1 of 2,4-D amine (Weedar® 64, Nufarm Agricultural Products, Alsip, IL) 24 h before malathion/PBO applications followed by the herbicide applications. The experiment was repeated once. Plants were treated with a track sprayer with the configuration described earlier. Plant mortality was recorded at 21 DAT. To assess the effect of additives to the herbicide on mortality rate, a chi-square test was conducted using PROC FREQ in SAS.
Results and discussion
Resistance to imazamox and mesosulfuron
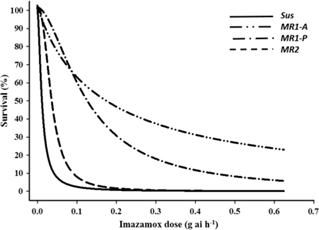
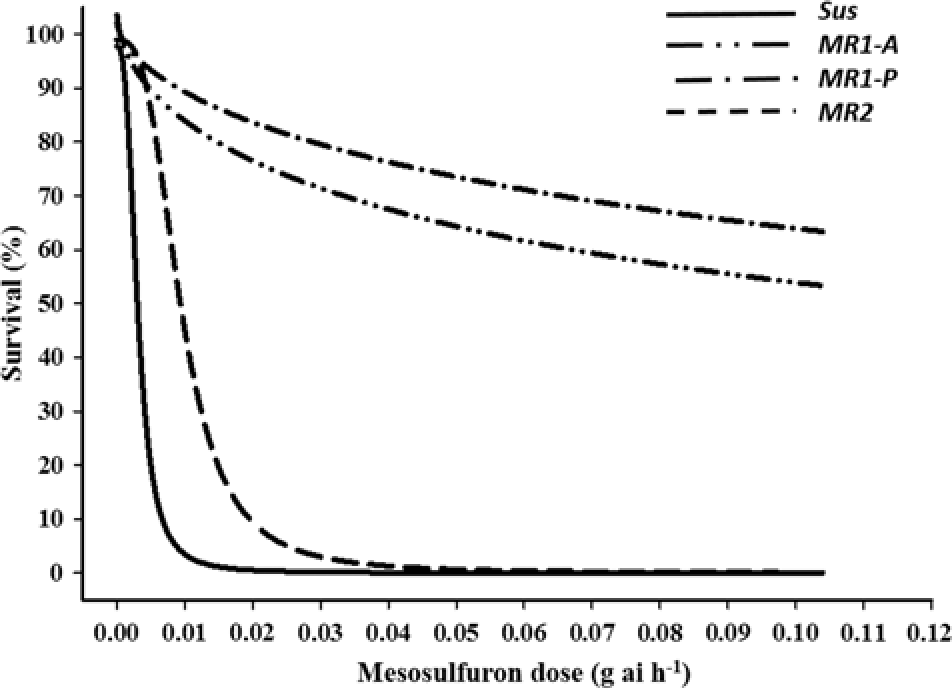
Both imazamox and mesosulfuron controlled 100% of the Sus individuals at the labeled field rates (Figures 1 and 2, respectively). In contrast, the MR subpopulations were resistant to both herbicides but differed significantly in resistance level. LD50 values for the MR1-A, MR1-P, MR2, and Sus plants were 0.19, 0.144, 0.04, and 0.005 g ha−1 of imazamox, respectively. R/S ratios, calculated from the LD50 values, indicated that the MR1-A, MR1-P, and MR2 plants were 38-, 29-, and 8-fold less sensitive to imazamox, respectively, than the Sus plants. LD50 values for the MR1-A, MR1-P, MR2, and Sus plants were 0.18, 0.319, 0.017, and 0.005 g ha−1 of mesosulfuron, respectively, representing a 36-, 64-, and 3-fold resistance to mesosulfuron for the MR plants, correspondingly, compared with the Sus plants. Resistance to imazamox and mesosulfuron, either individually or in combination, has been confirmed in several L. perenne ssp. multiflorum populations across the United States and in Denmark and Italy (Heap Reference Heap2018). For example, an accession from Arkansas was resistant to both imazamox and mesosulfuron (Kuk and Burgos Reference Kuk and Burgos2007), whereas a different mesosulfuron-resistant accession was controlled by imazamox (Kuk et al. Reference Kuk, Burgos and Scott2008). In a herbicide-resistance survey conducted across 75 fields in the Palouse region encompassing parts of eastern Washington and northern Idaho, 55% and 34% of L. perenne ssp. multiflorum populations were deemed to be resistant or developing resistance to imazamox and mesosulfuron, respectively (Rauch et al. Reference Rauch, Thill, Gersdorf and Price2010). Furthermore, two L. perenne ssp. multiflorum populations from winter wheat (Triticum aestivum L.) fields in Oregon were confirmed to be resistant to mesosulfuron (Liu et al. Reference Liu, Hulting and Mallory-Smith2016) and a biotype from Texas was 24-fold resistant to mesosulfuron (Ellis et al. Reference Ellis, Morgan and Mueller2008).
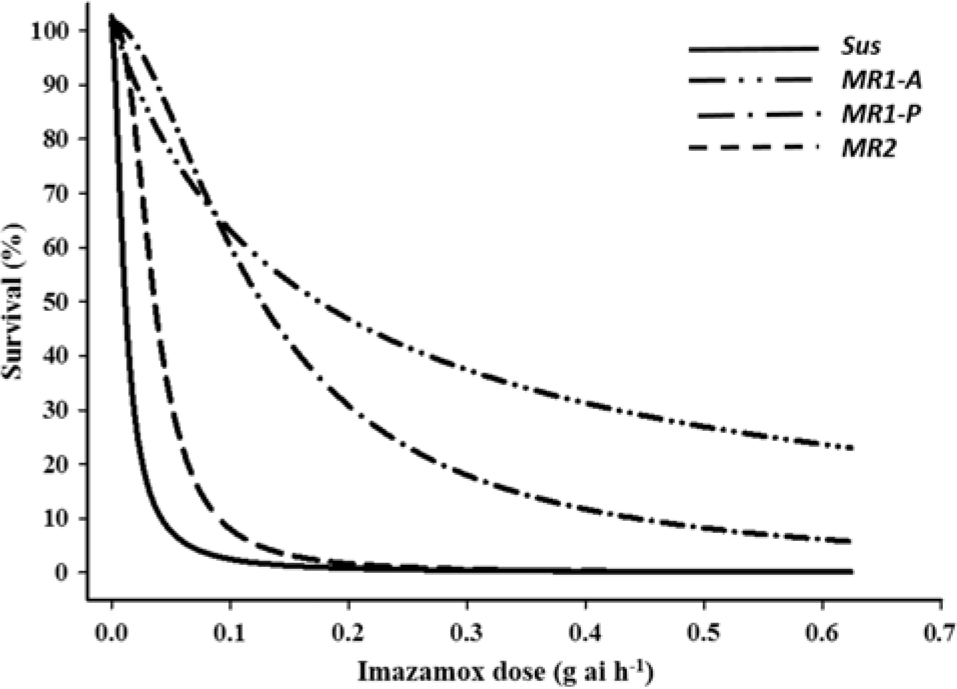
Figure 1. Imazamox dose response on survival of Lolium perenne ssp. multiflorum subpopulations (Sus, MR1-A, MR1-P, MR2) at 21 d after treatment.
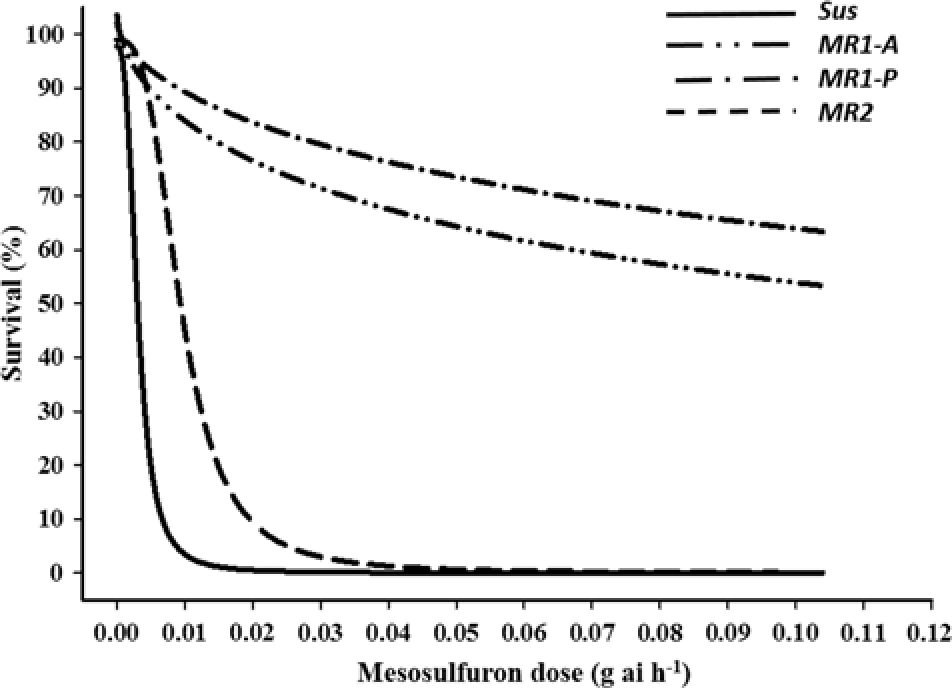
Figure 2. Mesosulfuron-methyl dose response on survival of Lolium perenne ssp. multiflorum subpopulations (Sus, MR1-A, MR1-P, MR2) at 21 d after treatment.
Cross- and multiple resistance
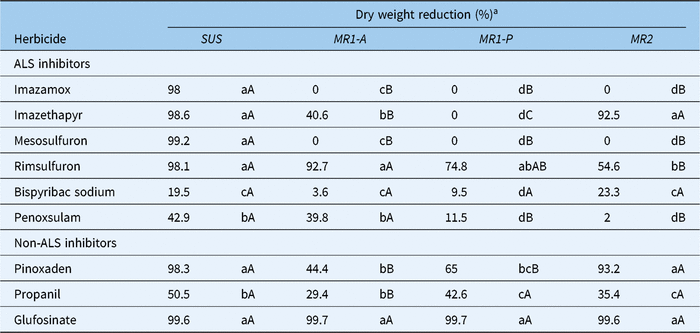
Aboveground biomass dry weight reduction, in response to treatment with ALS-inhibiting and several non–ALS inhibiting herbicides (Table 1), differed markedly among populations (Table 3). Imazethapyr reduced the aboveground biomass of MR1-A (40.6%) plants to a significantly lesser degree than MR2 (92.5%) and Sus (98.6%) but failed to have any impact on MR1-P (0%) plants. Interestingly, the response of plants to rimsulfuron differed from that for imazethapyr. At the labeled field rate, rimsulfuron resulted in only 54.6% and 74.8% aboveground biomass dry weight reduction of MR2 and MR1-P plants in contrast to 92.7% and 98.1% for MR1-A and Sus individuals, respectively. Among the non-ALS inhibitors, shoot dry weight reduction was similar for pinoxaden-treated plants from the MR1-A and MR1-P subpopulations, but significantly less than for MR2 and Sus (Table 3). Pinoxaden only controlled Sus (98.3%) and MR2 (93.2%) plants reasonably well, whereas glufosinate reduced the shoot dry weight of plants from all four populations by nearly 100%. Bispyribac and penoxulam and the non-ALS inhibitor propanil were also evaluated, but all three herbicides were ineffective on plants from all four populations (Table 3), explaining lack of L. perenne ssp. multiflorum being listed on their respective labels.
Table 3. Percentage reduction of aboveground dry weight biomass of Lolium perenne ssp. multiflorum (Sus, MR1-A, MR1-P, MR2) in response to treatment with ALS-inhibiting and non–ALS inhibiting herbicides
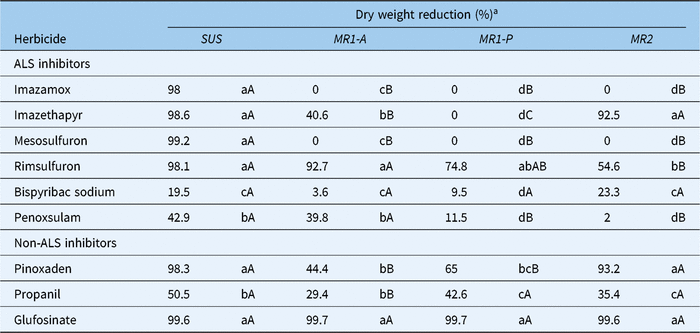
a Means for each accession within a column followed by the same lowercase letters and means for each herbicide within a row followed by the same uppercase letters are not significantly different according to Fisher’s protected LSD test (α = 0.05).
In summary, MR1-A and MR1-P, which are highly resistant to imazamox and mesosulfuron are cross-resistant to imazethapyr, and MR1-P is also cross-resistant to rimsulfuron. MR1-A and MR1-P are also multiple resistant to pinoxaden. In contrast, MR2, which has a much lower level of resistance to imazamox and mesosulfuron is only cross-resistant to rimsulfuron (Table 3).
ALS enzyme assay
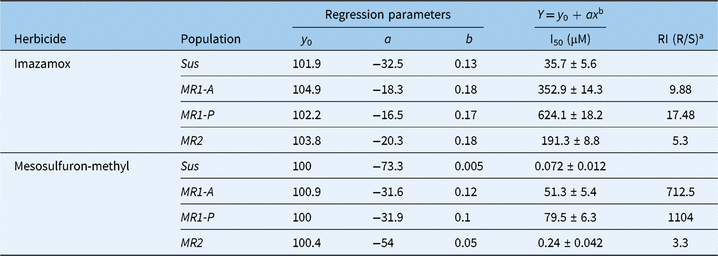
The ALS activity in the absence of herbicide of the four populations ranked as Sus > MR2 > MR1-A > MR1-P (unpublished data). I50 values for Sus, MR2, MR1-A (Table 4), and MR1-P populations were 0.072, 0.24, 51.3, and 79.5 µM, respectively, thereby indicating that the ALS enzyme from MR2, MR1-A, and MR1-P plants was 3-, 713-, and 1,104-fold, correspondingly, more resistant to mesosulfuron than the ALS from Sus plants. Kuk and Burgos (Reference Kuk and Burgos2007) reported a 7-fold difference in sensitivity of ALS to mesosulfuron in resistant and susceptible accessions from Arkansas. In general, a high level of resistance based on ALS enzyme inhibition by ALS-inhibiting herbicides indicates the presence of one or more point mutations resulting in amino acid substitutions in the ALS enzyme of resistant plants (Wright et al. Reference Wright, Bascomb, Sturner and Penner1998).
Table 4. Herbicide concentrations required for 50% inhibition (I50) of the ALS enzyme (± standard error of mean) of Lolium perenne ssp. multiflorum subpopulations (Sus, MR1-A, MR1-P, MR2) and the corresponding regression model parameters

a RI, resistance index (R/S).
ALS Sanger sequencing
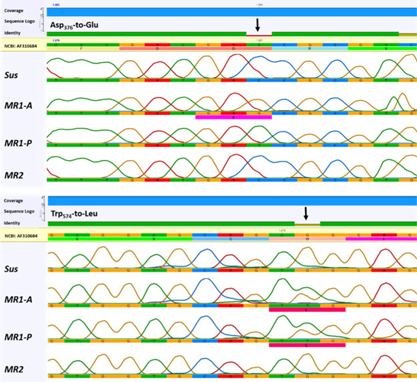
Partial ALS sequences of Sus, MR1-A, MR1-P, and MR2 plants (NCBI GenBank accession numbers: Sus = MH165308, MR1-A = MH165309, MR1-P = MH165310, and MR2 = MH165311) spanning eight codons where point mutations are known to confer resistance to ALS-inhibiting herbicides (Tranel et al. Reference Tranel, Wright and Heap2018) were amplified and sequenced. Alignment of resistant plant sequences revealed a missense single-nucleotide polymorphism resulting in a Trp-574-Leu substitution in MR1-A and MR1-P plants, heterozygous in both, that was not detected in MR2 plants (Figure 3). The Trp-574-Leu substitution is known to cause a high level of resistance to ALS inhibitors in L. perenne ssp. multiflorum and L. rigidum (Liu et al. Reference Liu, Hulting and Mallory-Smith2014; Tan et al. Reference Tan, Preston and Wang2007; Yu et al. Reference Yu, Han and Powles2008). An additional homozygous substitution, Asp-376-Glu, was identified in 75% of MR1-A plants, but not in MR1-P or MR2 plants. Recently, the Asp-376-Glu mutation was reported in an L. perenne population from France (Menegat et al. Reference Menegat, Bailly, Aponte, Heinrich, Sievernich and Gerhards2016). Congruently, multiple (six) resistance-conferring ALS mutations in an L. rigidum population (Yu et al. Reference Yu, Han and Powles2008) and double mutations in the ALS gene of imazethapyr- and chlorsulfuron-resistant sugar beet (Beta vulgaris L.) lines (Wright et al. Reference Wright, Bascomb, Sturner and Penner1998) have previously been reported.
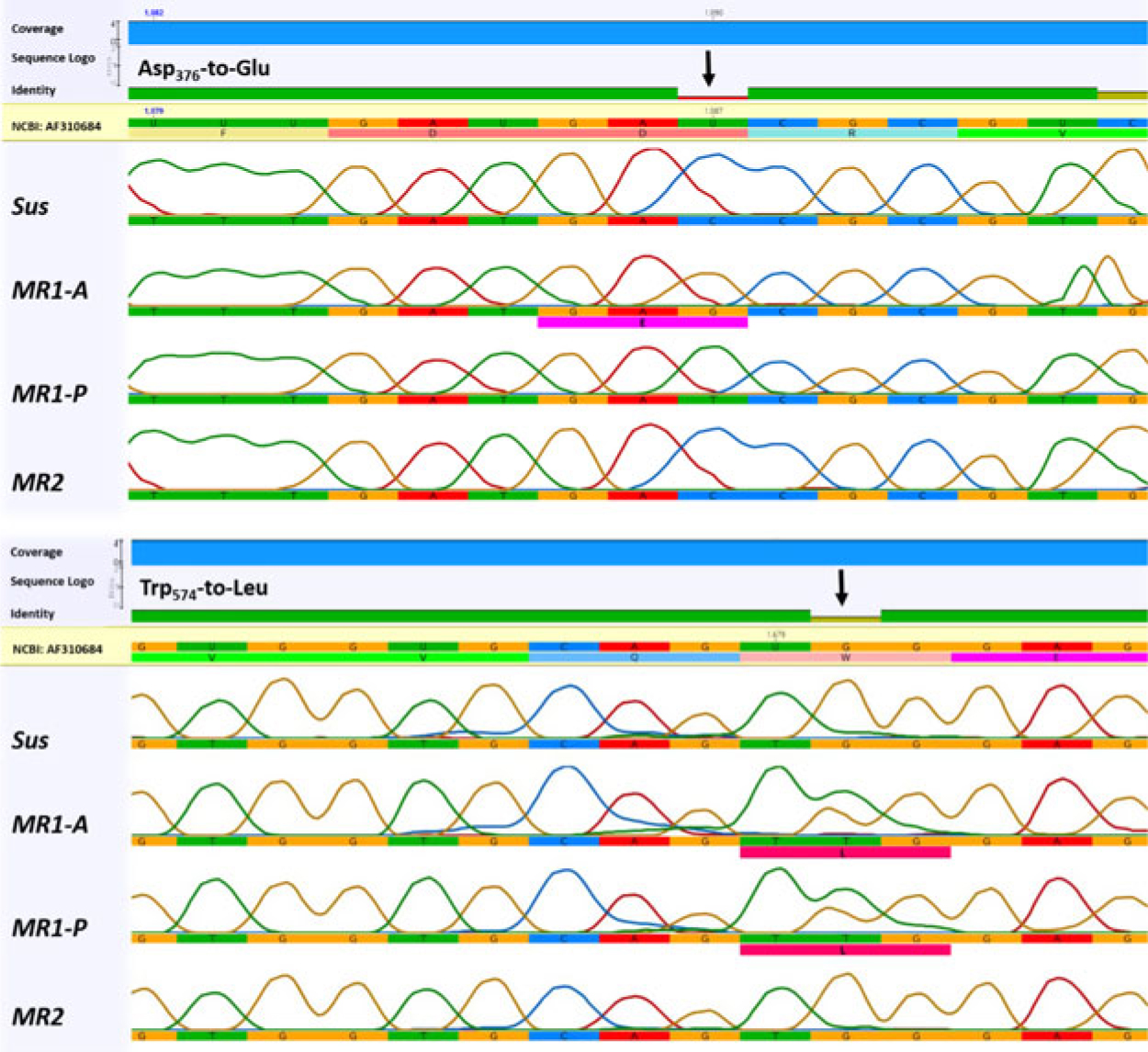
Figure 3. Comparison of ALS gene sequences in the region encompassing point mutations known to confer resistance to ALS-inhibiting herbicides in Lolium perenne ssp. multiflorum subpopulations (Sus, MR1-A, MR1-P, MR2) from California. The black arrows indicates the mutant codons.
CYP inhibition by malathion and PBO
Addition of malathion or PBO to mesosulfuron, irrespective of time of pretreatment, did not alter the efficacy of mesosulfuron on MR2 plants (unpublished data). In addition, presence of 2,4-D had no effect on the response of MR2 and SUS plants to mesosulfuron (unpublished data). These results suggest that the mechanism of resistance to mesosulfuron in plants from the MR2 subpopulation may not be based on metabolism of the herbicide. However, it is also possible that herbicide metabolism could be affected by CYP isozymes/enzymes that are not inhibited by malathion or PBO or that an herbicide metabolism pathway that does not involve CYP genes, such as glutathione-S-transferases, could be involved. CYP hemoproteins can also be inhibited selectively by various derivatives of triazole, imidazole, and pyrimidine (Donaldson and Luster Reference Donaldson and Luster1991). Additional research investigating the non–target site mechanism underlying the resistance of MR2 plants to ALS-inhibiting herbicides is needed.
In summary, our results confirm resistance to imazamox and mesosulfuron, two ALS-inhibiting herbicides, in MR1-A, MR1-P, and MR2 L. perenne ssp. multiflorum subpopulations from California, which have all previously been shown to have multiple resistance to glyphosate, paraquat, and ACCase-inhibiting herbicides (Tehranchian et al. Reference Tehranchian, Nandula, Jugulam, Putta and Jasieniuk2018). The levels of resistance to imazamox and mesosulfuron were several fold higher in the MR1-A and MR1-P subpopulations than in the MR2 based on whole-plant dose–response experiments and studies of ALS enzyme inhibition by the two herbicides. Moreover, plants from the subpopulations with the highest resistance level (MR1-A and MR1-P) revealed the Trp-574-Leu mutation, as observed in previous studies of biotypes highly resistant to ALS-inhibiting herbicides (e.g., Tranel and Wright Reference Tranel and Wright2002). An additional mutation, Asp-376-Glu, contributed to the resistance trait in MR1-A plants. Cross-resistance to other ALS inhibitors varied among the three multiple resistant L. perenne ssp. multiflorum. subpopulations. Pinoxaden was only effective on MR2 plants, while glufosinate controlled all three multiple resistant subpopulations. Treatment with CYP inhibitors failed to increase the susceptibility of the MR2 plants to mesosulfuron, making our results on herbicide metabolism inconclusive. It is plausible that other CYP enzymes not inhibited by the CYP inhibitors used in this research could play a role. Further investigations are warranted to determine the role of metabolism of mesosulfuron and imazamox as a non–target site resistance mechanism(s) in MR2 plants. For example, screening additional CYP inhibitors or direct investigation of the fate of mesosulfuron and imazamox in the MR2 plants could provide more conclusive evidence of absence or presence of a metabolic mechanism of resistance. Non–target site mechanisms, such as herbicide metabolism, can potentially render ineffective yet-to-be commercialized herbicides with unique modes of action. Meanwhile, prudent use of glufosinate is needed to decrease selection pressure for the evolution of resistance to this herbicide in these multiple resistant L. perenne ssp. multiflorum populations in northern California.
Author ORCIDs
Parsa Tehranchian https://orcid.org/0000-0001-9885-0182; Vijay K. Nandula https://orcid.org/0000-0001-5618-7634; Maor Matzrafi https://orcid.org/0000-0002-4867-0850; Marie Jasieniuk https://orcid.org/0000-0003-0388-6786.
Acknowledgments
This research was funded by USDA-NIFA-AFRI Award No. 2015-67013-22949, and MJ was supported by USDA National Institute of Food and Agriculture Hatch Project 1002110. No conflicts of interest have been declared. The authors would also like to thank Chad Fautt for greenhouse and laboratory assistance and Hannah Clifton and Kevin Roberts for greenhouse and environmentally controlled chamber maintenance.