Introduction
The genus Tremella Pers. (Tremellomycetes, Basidiomycota, Fungi) includes predominantly mycoparasitic species, growing on a wide range of basidiomycete and ascomycete fungi (Chen Reference Chen1998; Kirk et al. Reference Kirk, Cannon, Minter and Stalpers2008). Each particular Tremella species is, however, highly host-specific, frequently being confined to a single fungal genus or species. Many taxa form conspicuous gelatinous basidiocarps and are well known representatives of ‘jelly fungi’, such as Tremella mesenterica or Tremella foliacea. Lichenicolous species are among the most poorly known representatives of the genus. Fifty-one Tremella species have been described so far, growing exclusively on lichenized fungi (Diederich Reference Diederich1986, Reference Diederich1996, Reference Diederich2003, Reference Diederich2007; Sérusiaux et al. Reference Sérusiaux, Diederich, Ertz and van den Boom2003; Zamora et al. Reference Zamora, Pérez-Ortega and Rico2011), although five of these have not yet been formally named (Diederich Reference Diederich1996, Reference Diederich2007). The lichenicolous Tremella species often induce the formation of relatively conspicuous galls on their hosts, either on the thallus or on the hymenium of the ascocarps. Some intrahymenial taxa do not produce any external symptoms, at least not in early stages of growth (Diederich Reference Diederich1996; Zamora et al. Reference Zamora, Pérez-Ortega and Rico2011). The phylogenetic position of the lichen-inhabiting representatives had never been tested by molecular methods until the work by Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011), who confirmed that they were nested within the genus Tremella. However, Tremella as currently circumscribed is strongly polyphyletic (Chen Reference Chen1998; Fell et al. Reference Fell, Boekhout, Fonseca, Scorzetti and Statzell-Tallman2000; Scorzetti et al. Reference Scorzetti, Fell, Fonseca and Statzell-Tallman2002; Boekhout et al. Reference Boekhout, Fonseca, Sampaio, Bandoni, Fell, Kwon-Chung, Kurtzman, Fell and Boekhout2011; Millanes et al. Reference Millanes, Diederich, Ekman and Wedin2011), and both the genus and the family Tremellaceae are in need of a deep taxonomic revision (Boekhout et al. Reference Boekhout, Fonseca, Sampaio, Bandoni, Fell, Kwon-Chung, Kurtzman, Fell and Boekhout2011; Millanes et al. Reference Millanes, Diederich, Ekman and Wedin2011). In their study of Tremellomycetes, Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011) found that the lichenicolous lifestyle was not restricted to a single clade, and they distinguished three monophyletic groups including mostly lichenicolous species.
It has been suggested that the diversity of lichen-inhabiting fungi is, in general, probably underestimated (Lawrey & Diederich Reference Lawrey and Diederich2003; Ihlen & Wedin Reference Ihlen and Wedin2008) and this is also the case in Tremella in particular, where the number of new descriptions of lichen-inhabiting taxa is certainly expected to increase in the future (P. Diederich, pers. obs.; Zamora et al. Reference Zamora, Pérez-Ortega and Rico2011). Moreover, the frequency of new lichenicolous Tremella records reported in recent years suggests that many of the species already described might have been largely overlooked in previous field surveys, and that the distribution of many species is possibly wider than currently considered (Diederich Reference Diederich2003; Sérusiaux et al. Reference Sérusiaux, Diederich, Ertz and van den Boom2003; Ertz & Diederich Reference Ertz and Diederich2008; Kukwa & Jabłonska Reference Kukwa and Jabłońska2008; Pippola & Kotiranta Reference Pippola and Kotiranta2008; Puolasmaa et al. Reference Puolasmaa, Pippola, Huhtinen, Hyvärinen and Stenroos2008; Westberg et al. Reference Westberg, Millanes and Wedin2008; Svensson & Westberg Reference Svensson and Westberg2010).
During fieldwork on the island of Runmarö in the Stockholm archipelago (Sweden), the second author collected a specimen of the lichen Diploschistes scruposus showing unusual dark galls on the thallus. Further microscopic observations revealed the presence of basidia and basidiospores of the Tremella type inside these galls. The same author later found two other specimens growing on Diploschistes scruposus in Sweden, and a fourth sample, growing on Diploschistes muscorum, had been collected previously by Roger Rosentreter in the USA and sent to Paul Diederich. No other Tremella species had been previously recorded on these hosts, and we conclude that the four specimens correspond to a new species of lichen-inhabiting Tremella, which is described here as Tremella diploschistina sp. nov., on the basis of morphological and molecular studies. Its phylogenetic relationship to other species in the genus is also investigated by molecular methods.
Material and Methods
Morphological studies
Macromorphological traits were observed using an Olympus SZX16 dissecting microscope. Microscopic structures were studied using hand-cut sections stained with Phloxin (1% in water) after pre-treatment with KOH (5%), following the methods of Diederich (Reference Diederich1996), and observed with an Olympus CX40 microscope. Drawings were performed using a drawing tube and by direct observation. Micrographs were taken using an Olympus BX53 microscope fitted with differential interference contrast (DIC) and an Olympus DP11 camera. Mycological terminology follows Diederich (Reference Diederich1996) and Kirk et al. (Reference Kirk, Cannon, Minter and Stalpers2008). The apiculus was not included in basidiospore measurements. Sizes in parentheses represent minimum and maximum observed values. When the number of observations is less than 30, it is indicated in brackets.
Molecular studies
Choice of additional taxa and outgroup
In addition to the three specimens studied, 18 specimens representing 10 Tremella species were included in the molecular study (Table 1). The sampling included the type of the genus Tremella (T. mesenterica), terminals of the Fuciformis and Foliacea groups distinguished by Chen (Reference Chen1998) and terminals representing three groups of lichenicolous species distinguished by Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011). We included two species of Filobasidiales, viz., Filobasidium floriforme and F. uniguttulatum, as outgroup.
Table 1. Sequences newly produced (bold) or downloaded from GenBank, with specimen data or culture references.

* Type specimen of the new species.
Species names, voucher information, and GenBank accession numbers are given in Table 1.
DNA extraction and amplification
DNA was extracted directly from the three specimens examined (Table 1). The outer surface of the selected galls, in which most of the tremellalean hyphae and hymenial components are located, was sectioned and separated with a scalpel, in order to minimize the lichen tissue in the DNA extraction. Total DNA was extracted using the Qiagen DNeasy Plant MiniKit, according to the manufacturer's instructions.
For PCR amplification we used general fungal primers in combination with primers designed to selectively amplify the DNA from tremellalean fungi (Millanes et al. Reference Millanes, Diederich, Ekman and Wedin2011). The primers ITS1F (Gardes & Bruns Reference Gardes and Bruns1993), BasidLSU3-3 and BasidLSU1-5 (Millanes et al. Reference Millanes, Diederich, Ekman and Wedin2011), and LR5 (Vilgalys & Hester Reference Vilgalys and Hester1990) were used to amplify the internal transcribed spacer I, the 5.8 rDNA gene, the internal transcribed spacer II and a fragment of approximately 1000 bp in the nLSU rDNA gene.
PCR amplifications were performed using Illustra™ Hot Start PCR beads, according to the manufacturer's instructions, with the following settings: for the primer pair ITS1F/BasidLSU3-3, we used initial denaturing at 95°C for 3 min, four cycles (95°C for 40 s, 53°C for 40 s and 72°C for 90 s), four cycles (95°C for 30 s, 50°C for 30 s and 72°C for 90 s), and finally 32 cycles (95°C for 30 s, 47°C for 30 s and 72°C for 90 s) with a final extension at 72°C for 480 s. For the primer pair BasidLSU1-5/LR5 we used initial denaturing at 95°C for 3 min, four cycles (95°C for 40 s, 56°C for 40 s and 72°C for 90 s), four cycles (95°C for 30 s, 53°C for 30 s and 72°C for 90 s) and finally 32 cycles (95°C for 30 s, 50°C for 30 s and 72°C for 90 s) with a final extension at 72°C for 420 s. Before sequencing, the PCR products were purified using the PCR-M® Clean-up System of Viogene or the enzymatic method Exo-sap-IT© provided by USB Corporation.
Sequence alignment and phylogenetic analyses
Sequences were aligned using the Q-INS-i algorithm (Katoh & Toh Reference Katoh and Toh2008a) of the multiple sequence alignment software MAFFT version 6.611 (Katoh et al. Reference Katoh, Misawa, Kuma and Miyata2002; Katoh & Toh Reference Katoh and Toh2008b), following Wedin et al. (Reference Wedin, Wiklund, Jørgensen and Ekman2009), but aligning sequences in a single step. Major insertions and ambiguous regions were identified and eliminated with Gblocks version 0.91b (Castresana Reference Castresana2000).
Dataset congruence was assessed manually by analyzing the datasets separately by parsimony bootstrapping. Conflict among clades was considered as significant if a significantly supported clade (bootstrap support ≥ 70%; Hillis & Bull Reference Hillis and Bull1993) for one marker was contradicted with significant support by another. No incongruence was found and the data were concatenated into a single dataset.
Maximum parsimony and parsimony bootstrap analyses were performed for the combined dataset using PAUP* 4.0b10 (Swofford Reference Swofford2002) with the following settings: gaps were treated as ‘missing data’, 1000 random addition sequence replicates, TBR branch swapping, steepest descent off, collapse branches if minimum length is 0, MulTrees on. Bootstrap (Felsenstein Reference Felsenstein1985): heuristic search settings identical to the above analysis but with 10 random addition replicates; bootstrap settings: 1000 bootstrap replicates, full heuristic search, retain groups with frequency >50%. Parsimony-uninformative characters were excluded from these analyses.
Bayesian inference of phylogeny (Huelsenbeck et al. Reference Huelsenbeck, Ronquist, Nielsen and Bollback2001) was carried out by Markov Chain Monte Carlo (MCMC) sampling as implemented in the software MrBayes 3.1.2 (Huelsenbeck & Ronquist Reference Huelsenbeck and Ronquist2001). Likelihood models were selected for each of the three gene regions using the Bayesian Information Criterion (BIC) as implemented in jModeltest (Posada Reference Posada2008). We used full likelihood optimization and selected only among the 24 models implemented in MrBayes. Following this scheme, a SYM+Γ model was selected for the ITS, and a SYM+I+Γ for the nuclear LSU rDNA. The combined analysis treated the two gene regions as separate partitions with topology linked across partitions but separate model parameter values and proportional rates across partitions. The number of discrete gamma categories was kept at default four. Bayesian prior distributions included treating all tree topologies as equally likely, a uniform (0, 50) distribution for the gamma shape parameter, a uniform (0, 1) distribution for the proportion of invariable sites, and a flat (1, 1, 1, 1, 1, 1) Dirichlet for the rate matrix. For the combined dataset, two parallel runs were performed, each with four chains, three of which were incrementally heated with a temperature of 0·15. The analysis was diagnosed for convergence every 100 000 generations, measured as the average standard deviation of splits across runs in the last half of the analysis. Every 100th tree was saved. The first half of the run was discarded as burn-in.
The Species
Tremella diploschistina Millanes, M. Westb., Wedin & Diederich sp. nov
MycoBank No: MB563327
Basidiomata lichenicola in thallo Diploschistis, gallas superficiales, luteas, atrobrunneas, vel atras, convexas, basim non constrictas 0·3–0·9 mm in diam. efficientia. Hymenium hyphidiis tumidis, elongatis, septatis, ramosis, 3·5–5 µm in diam. Basidia 2-cellularia, septo longitudinali, obliquo vel transversali (13–)14–30 (–34) × 8–14 µm. Basidiosporae 7–9 × (5–)6–9 µm. Conidia ignota.
Typus: Sweden, Uppland, Djurö par., Runmarö, Norestranden NE of Nore, 59°16′43″N, 18°47′47″E, 30 June 2009, M. Westberg & T. Berglund 09-400 (S—holotypus).
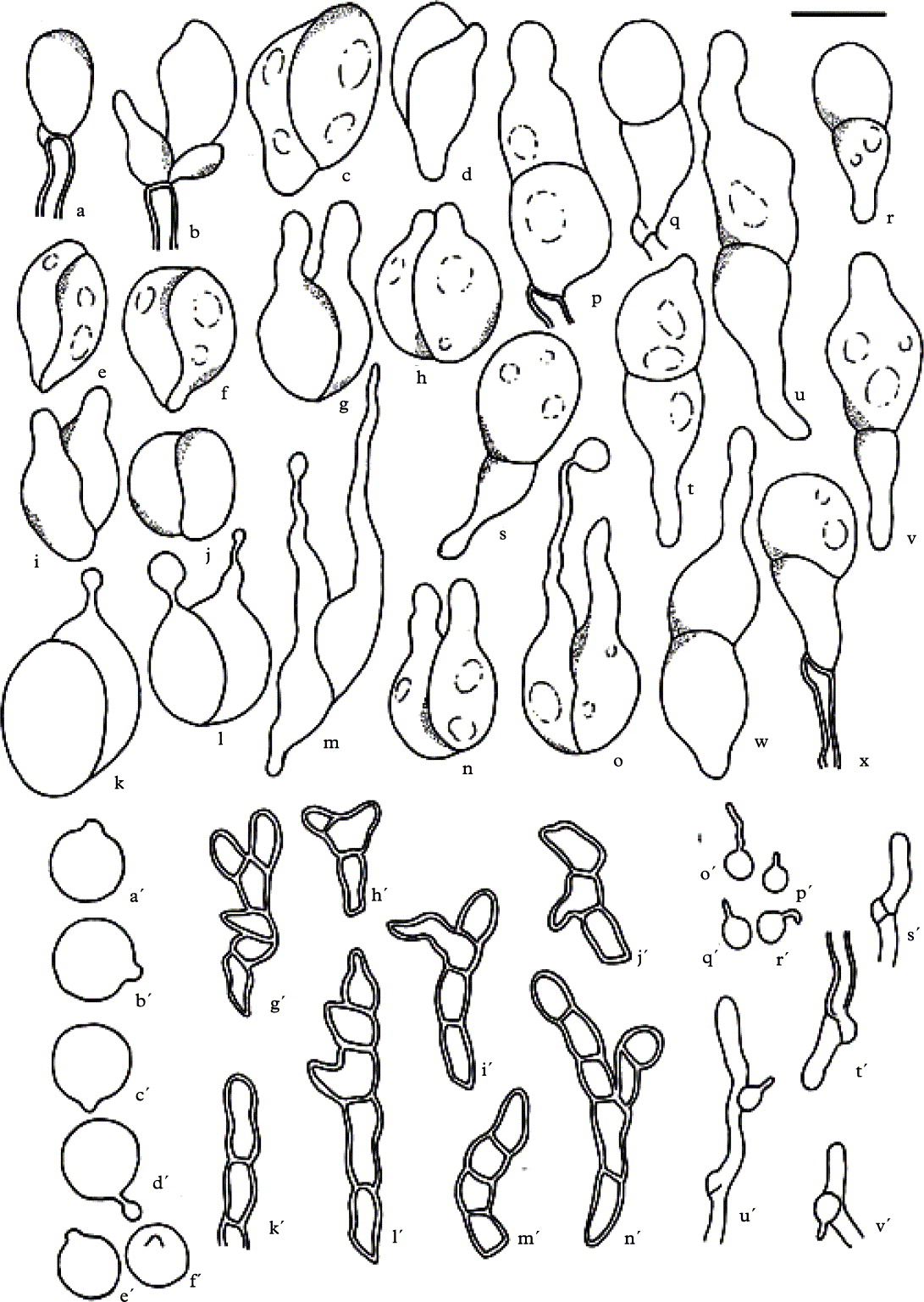
Fig. 1. Tremella diploschistina. a–x, basidia; a'–f', basidiospores; g'–n', hyphidia; o'–r', haustorial branches; s' & t', hyphae with clamps; u' & v', hyphae with haustorial cells; a, d, l–m, s, v–w, e'–f', i'–j', m'–n', and o'–r' (IMI 35641); j–k, n–q, t–u, e'–f', i'–j', m'–n' (M. Westberg 09-452 (S)); b–c, e–i, r, x, a'–b', k'–l' and s' (holotype). Scale = 10 µm.

Fig. 2. Tremella diploschistina. A, habit; B–E, basidia showing different septation pattern; F, basidiospore. A & B (holotype); C & D (IMI 35641); F (M. Westberg 09-452 (S)). Scales: A = 1 mm; B–F = 10 µm.
Basidiomata waxy, inducing the formation of galls on the thallus surface (Fig. 2A). Galls pale yellow, dark brown, or black, at first regularly convex to subglobose, 0·3–0·9 mm diam., often forming groups measuring up to 3 mm diam. Context hyphae thin-walled, often with clamp connections, 1·5–2·5 µm diam. (Fig. 1s'–v'); haustorial branches frequent, mother cells spherical to subspherical, 3–4 × 3–4 µm, haustorial filament 1 µm diam., up to 8 µm long (Fig. 1o'–r'). Hymenium hyaline, containing numerous probasidia; hyphidia present, thick-walled, with numerous septa and ramifications, 3·5–5·0 µm diam. (Fig. 1g'–n'), sometimes swollen with thin walls, then up to 6 µm diam. (n = 11). Probasidial initials clavate, proliferations occurring through the basal clamp (Fig. 1a & b). Basidia, when mature, 2-celled, with one transverse, oblique or longitudinal septum. The three types of basidium septation are often found within the same gall. When transverse, constricted at the septum, the lower cell with an attenuated stalk-like base, often longer than the upper cell, (13–)14–30(–34) × 8–14 µm (incl. stalk-like base; excl. epibasidia); lower part of the stalk-like base 2–4 µm diam.; epibasidia subcylindrical, at least up to 30 µm long, 2–4 µm diam. (Figs 1c–x and 2B–D). Basidiospores ellipsoid to subspherical, c. 7–9 × (5–)6–9 µm (n = 21) with a distinct apiculus, c. 1 µm diam. (Figs 1a'–f' & 2F).
Anamorph not observed.
Hosts. On the thallus of Diploschistes. Swedish samples grew on Diploschistes scruposus and the USA sample grew on D. muscorum.
Distribution and ecology. Known from northern Europe (Sweden) where it grows on exposed siliceous rocks, predominantly lake- or seashore rocks, but also in forested areas, and from North America (USA, Idaho) where it occurs in an Artemisia tridentata and Agropyron spicatum habitat on sandy loam soil.
Additional specimens examined. Sweden: Dalsland: Skållerud par., Lake Östebosjön, W side and near S tip of island Hinnön, 58°49′4″N, 12°28′51″E, 2009, M. Westberg 09-452 (S). Uppland: Djurö par., Runmarö, Norestranden NE of Nore, 59°16′43″N, 18°47′48″E, 14 v 2010, M. Westberg (hb. Diederich). Södermanland: Nacka par., SE part of Nyckelviken Nature Reserve, 59°19′6″N, 18°11′38″E, 14 v 2011, M. Westberg (S).—USA: Idaho: S of Emett, Old Freeze Out Hill, north facing hillside, T6N R1W, 1991, R. Rosentreter 6836 (IMI 35641, hb. Rosentreter).
Phylogenetic Results
We generated 6 new sequences (3 ITS and 3 nLSU rDNA), which were aligned together with sequences already available in GenBank (Table 1). Two data matrices were produced, one including ITS and one including nLSU rDNA. The three ITS sequences of Tremella diploschistina differed in two nucleotides among them, and 10 ITS positions differed from the rest of ITS Tremella sequences studied. Only 1 position differed among the three T. diploschistina nLSU sequences, and 16 positions in the nLSU were different from the other Tremella species studied.
The combined matrix contained 1212 characters (ITS: 1-300; nLSU: 301-1212), from which 348 unambiguously aligned parsimony informative sites were used inthe parsimony analysis. Our maximum parsimony analysis resulted in 2 most parsimonious trees of 694 steps, with CI = 0·622 and RI = 0·773.
When the Bayesian analysis halted after 200 000 generations, the average standard deviation of split frequencies across runs was 0·004, which indicates that the two runs have converged (<0·01). A majority rule consensus tree was constructed from the 2000 trees of the stationary tree sample.
The three specimens of T. diploschistina formed a single clade, supported both by parsimony bootstrap (100%) and Bayesian Posterior Probabilities (1.0) (Fig. 3). The Tremella species included in our analysis also formed a supported clade (parsimony bootstrap = 100% and BPP = 1·0), including the clade formed by the three specimens of T. diploschistina.

Fig. 3. Fifty per cent majority rule Bayesian consensus tree with average branch lengths from the combined analyses of ITS and nSSU datasets. PP values ≥ 0·95, obtained in the Bayesian analysis, are indicated over the branches, and bootstrap values ≥70%, obtained in the parsimony analysis, below the branches. Branch lengths are scaled to the expected number of nucleotide substitutions per site. A grey box encloses the clade containing the new species.
Discussion
Tremella diploschistina is distinguished from other known lichenicolous species growing on the host thallus with two-celled basidia with variable septation pattern, by the bigger size of basidia and basidiospores, the presence of hyphidia in the hymenium, and the different host-selection (Table 2). Two other species, T. psoromicola and T. stictae, form ramified hyphidia similar to those found in T. diploschistina, although the hyphidial walls are thicker in the latter species. Also, these two species grow on hosts of Peltigerales, and basidia of T. psoromicola do not show longitudinal septa. However, T. psoromicola was described from a single specimen, and we do not discard the possibility that longitudinal septa might be observed in additional collections. Other morphological traits such as basidium size and basidiospore size are within the ranges of T. diploschistina, suggesting a possible relationship to both species. Tremella stictae, however, has smaller basidia and basidiospores, and an asteroconidia-producing anamorph has been observed in this species, which is not known in T. diploschistina.
Table 2. Morphological and anatomical characters of Tremella diploschistina, compared to morphological characters of other Tremella species growing on the thallus of the host, and also bearing two celled basidia with variable septation pattern, and to morphological characters of other Tremella species growing on Graphidaceae. Data of all species except T. diploschistina are taken from Diederich (1996, 2007).

As species of Diploschistes are widely distributed and common, T. diploschistina is probably much more common and widespread than currently known. The genetic similitude between distantly collected specimens from the USA and Sweden, growing on different Diploschistes species (Fig. 3), is remarkable. Together with the morphological results, our molecular data further support the establishment of T. diploschistina as a well-delimited species, which appears clearly nested within Tremella in our molecular study, although the phylogenetic relationship with other Tremella species is not supported (Fig. 3). Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011) suggested that species in the Foliacea group might represent a distinct genus, if Tremella should be split in different taxa, but our new species is clearly not closely related to T. foliacea (Fig. 3). Preliminary analyses including T. diploschistina in the general phylogeny of the Tremellales did not support the phylogenetic relationship of the new species with any other taxa in the group (analysis based on the matrix from Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011); data not shown here).
Only two other lichenicolous Tremella species have been described so far on species of Graphidaceae sensu Mangold et al. (Reference Mangold, Martín, Lücking and Lumbsch2008) and Rivas-Plata & Lumbsch (Reference Rivas Plata and Lumbsch2011), viz., T. phaeographidis and T. phaeographinae (Diederich Reference Diederich1996). However, basidia of T. phaeographidis do not show transverse septa, basidiomata are flattened, pale to dark brown or reddish brown, basidia and basidiospores are smaller, and asteroconidia have been observed. Tremella phaeographinae forms basidiomata only in the hymenium of the host, which becomes reddish at maturity, the basidia can be 3-celled, the basidiospores are smaller, and blastoconidia have been observed. It would be interesting to test in the future whether, despite their morphological differences, the three Tremella species growing on Graphidaceae, (i.e. T. diploschistina, T. phaeographinae and T. phaeographidis) could be closely related. Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011) found that several lichenicolous species growing on Parmeliaceae (Biatoropsis usnearum, T. cetrariicola, T. coppinsii, T. everniae, and T. hypogymniae) were all nested within the same monophyletic group, although their micro- and macromorphology were clearly divergent.
It is interesting that Diploschistes is the only genus of the ‘Thelotrema clade’ of the Graphidaceae with a trebouxioid photobiont. Moreover, thalli of most Diploschistes species contain orcinol depsides, differing from most thelotremoid taxa, which usually have β-orcinol depsidones (Mangold et al. Reference Mangold, Elix, Lumbsch and McCarthy2009). Little if anything, however, is known on the factors influencing the high host-specificity observed in the lichenicolous Tremella species, and it has not been investigated whether the secondary lichen compounds could be involved in this specificity. Also, no kind of interaction between lichenicolous Tremella species and the photobiont of their lichenized host has ever been reported, and the interaction is considered to be exclusively mycoparasitic (Grube & de los Ríos Reference Grube and de los Ríos2001). Unfortunately, we could not amplify any DNA from the two Tremella species growing on other Graphidaceae, in order to test their possible relationship with T. diploschistina. Moreover, not all lichenicolous Tremella species described have been sequenced yet, and therefore future molecular studies adding more lichenicolous representatives might reveal the phylogenetic relationships of T. diploschistina with other lichen-inhabiting taxa in the Tremellales.
We thank David Hawksworth and Roger Rosentreter for sending us material for the present study and also the staff of the Molecular Systematic Laboratory at the Swedish Museum of Natural History in Stockholm (MSL, NRM) for their excellent technical support. This paper was financially supported by grants from the Swedish Research Council (VR 621-2009-537) to Mats Wedin, SYNTHESYS project SE-TAF-354, and Universidad Rey Juan Carlos (‘Estancias Breves’), Madrid, to Ana Millanes. Martin Westberg is supported by the Swedish Taxonomy Initiative (Svenska Artprojektet) administered by the Swedish Species Information Centre (ArtDatabanken).







