Introduction
Intestinal parasites such as soil-transmitted helminths (STH) are associated with significant morbidity in developing countries and are considered to be neglected pathogens (Pullan et al., Reference Pullan2014). It is estimated that 2 billion people are infected by STH and 54 million harbour Schistosoma mansoni worldwide (WHO, 2017, 2018). Both infections have been targeted by interventions supervised by the World Health Organization (WHO), which are based on mass administration of single-dose anthelmintics, such as albendazole. These interventions aim to control morbidity and parasitic burden and reduce transmission (WHO, 2017).
Kato–Katz is a (semi) quantitative stool examination technique described by Kato and Miura (Reference Kato and Miura1954) and modified by Katz et al. (Reference Katz, Chaves and Pellegrino1972). It aims to identify and quantify the number of eggs per gram (epg) of faeces and to estimate the intensity of infection by a given parasite. This technique, which is simple, inexpensive and field-applicable, is recommended by the WHO and has been widely used as a parasitological test for epidemiological surveys (WHO, 2015). Despite this, several studies have demonstrated a number of limitations, including (1) underestimation and low sensibility of prevalence when the parasitic load is low (Bärenbold et al., Reference Bärenbold2017; Lindholz et al., Reference Lindholz2018), (2) problems in detecting hookworm infections, as the stool must be prepared immediately (van Mens et al., Reference van Mens2013), and (3) the clarification step (i.e. glycerin) can render the eggs unrecognizable (Odongo-Aginya et al., Reference Odongo-Aginya2007). Its preparation can be carried out in the field and analysed by light microscopy, which requires only a very low-complexity infrastructure. The technique uses a small amount of previously manually homogenized fresh faecal matter (Montresor et al., Reference Montresor1998), which is applied on a screen to remove coarse particles, sized (c. 41.7 mg) on a perforated plate and compressed under a coverall of cellophane paper soaked in malachite green stain.
When applied to parasitology, molecular biology techniques can answer questions in the field of taxonomy, evolutionary genetics, molecular epidemiology and parasite load of infections (Demeler et al., Reference Demeler2013; O'Connell and Nutman, Reference O'Connell and Nutman2016; Hasegawa et al., Reference Hasegawa2017). They are also able to monitor the emergence of resistance to anthelmintics used on a large scale by the identification of single nucleotide polymorphisms in the nematode beta-tubulin gene (Diawara et al., Reference Diawara2013). These techniques may also be useful in the study of intraspecific genetic variability in the context of the development of STH vaccines (Stutzer et al., Reference Stutzer2018). In this context, we describe helminth DNA recovery, polymerase chain reaction (PCR) amplification and nucleotide sequencing directly from Kato–Katz thick smears prepared and read 7 years prior to molecular processing.
Materials and methods
This study used Kato–Katz slides (n = 124) prepared in 2011 during field work in the municipality of Santa Isabel do Rio Negro, Brazilian Amazon (0°28′S, 65°32′W), for STH diagnosis. The slides were prepared with fresh faeces using HelmTest (P&D CasaLab, Belo Horizonte, Minas Gerais, Brazil). Briefly, a previously homogenized fragment of the stool was passed through a metal mesh to remove coarse debris, and deposited in the orifice of a perforated plate. The stool cylinder molded by the plate was covered by a cellophane coverslip soaked in malachite green. The sample was compressed to spread the faeces under the coverslip, and examined under a light microscope. The slides were transported to the Laboratory of Epidemiology and Molecular Systematics in Rio de Janeiro (Oswaldo Cruz Foundation) and stored in a slide box at room temperature in a laboratory environment. After a few weeks, the parasite load for Ascaris sp. was evaluated. The result was multiplied by 24 to obtain the parasite load (eggs per gram of faeces, epg) according to the manufacturer's instructions. Reference values of classes of intensity are described in Montresor et al. (Reference Montresor1998).
In December 2017, the slides were reevaluated for the presence of parasite eggs (fig. 1a). The cellophane coverslip was removed and the remaining faecal material was scraped with a sterile scalpel. The faecal material was rehydrated (1 : 2 w/v) with 0.9% sterile saline for 48 hours at 4°C and macerated with a mini-pistil. DNA extraction was performed using a DNeasy Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
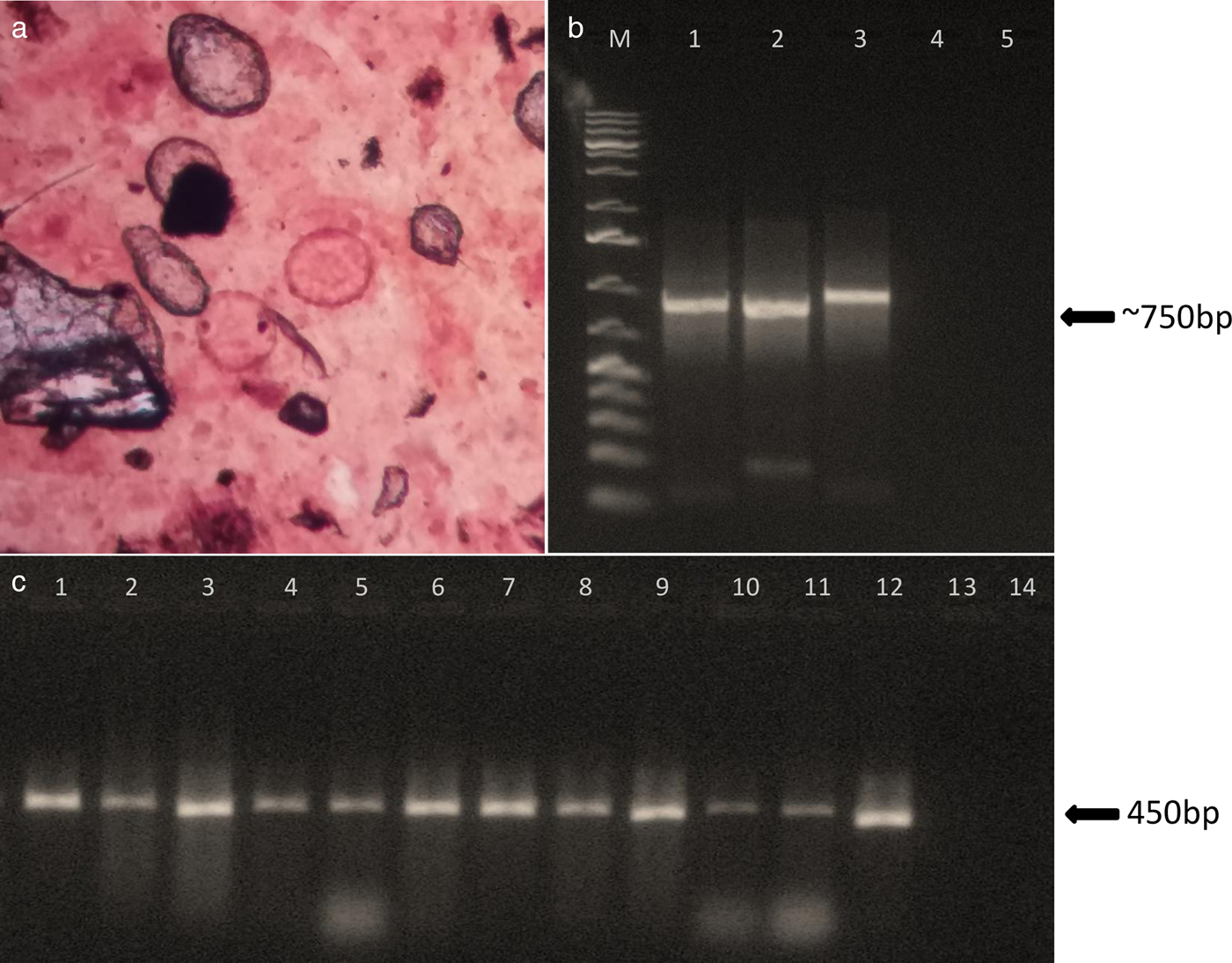
Fig. 1. Parasitological and molecular diagnosis of soil-transmitted helminths. (a) Ascaris sp. eggs. (b) Agarose gel electrophoresis 1.5% showing PCR amplification of Necator americanus ITS1-5.8S-ITS2 locus. Lane M: 1 Kb ladder; lanes 1, 2 and 3: positive samples; lane 4: negative sample; lane 5: negative PCR control. (c) Agarose gel electrophoresis 1.5% showing PCR amplification of Ascaris sp. cox1 locus. Lanes 1–12: positive samples; lane 13: negative sample; lane 14: negative PCR control.
PCR was performed using a Platinum Taq DNA Polymerase Kit (Invitrogen, Waltham, MA, USA), with a final volume of 25 μl. Mitochondrial or nuclear genetic targets are described in table 1. PCR conditions were as described by Peng et al. (Reference Peng2005) and Monti et al. (Reference Monti1998). The PCR products were electrophoresed in 1.5% Agarose High Resolution (Sigma-Aldrich, St. Louis, USA) in tris–acetate–EDTA gels stained with GelRed® (Biotium, Fremont, CA, USA). The sensitivity of the PCR technique was calculated using the following formula:
Table 1. Primer set used for amplification of soil-transmitted helminths.

cox1: cytochrome c oxidase subunit 1; ITS: internal transcribed spacer
To confirm the parasitic nature of the amplifications, nonspecific amplifications were excluded, molecular taxonomy was performed and PCR products were sequenced. The amplicons were purified using Illustra GFX PCR DNA and a Gel Band Purification Kit (GE HealthCare, Pittsburgh, PA, USA) and were sequenced using a BigDye Terminator v.3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) in an ABI 3730 Automated DNA Sequencer (Applied Biosystems). Bioedit v.7.2.5 (Hall, Reference Hall1999) was used to edit the sequences. The Basic Local Alignment Search Tool (BLAST, NCBI: https://www.ncbi.nlm.nih.gov/) was used to check similarity with parasite sequences. The GenBank accession numbers were MH665842–MH665844, MH674438–MH674442 and MH800218-MH800286.
Results and discussion
A total of 124 Kato–Katz thick smears were analysed. Of these, 93 (75%) were positive for Ascaris sp. and four (3.2%) were positive for hookworms. After a few weeks of collection in the field, the parasite load of Ascaris sp. only was assessed. The parasite load on the slides ranged from 96 to 54,000 (mean 8994) epg for Ascaris sp. Sixty percent (n = 56) of the slides showed moderate intensity infections, and 38.5% (n = 36) and 1.2% (n = 1) showed light and heavy intensity infections, respectively.
Ascaris sp. partial cox1 locus was amplified successfully in 84% (78/93) of the positive slides analysed (fig. 1c). The PCR products (423 bp) were sequenced in 74 samples and all were confirmed to be Ascaris lumbricoides or A. lumbricoides/A. suum when compared with sequences available in GenBank. PCR positivity in samples with different parasite loads was as follows: 89% (32/36) in samples with light infections, 80% (45/56) in samples with moderate infections, and 100% (1/1) in samples with heavy parasite loads. The sensitivity of the PCR technique was 84%. No correlation was found between PCR positivity and the intensity of infections. An explanation of the failure of amplification even with moderate parasite loads (between 5000 and 49,999 epg) could be the presence of PCR inhibitors, which we may consider intrinsic to DNA obtained from faecal samples. The purification of DNA samples or use of stool-specific kits may help to eliminate PCR inhibitors. All measures for non-contamination were taken, including the use of isolated rooms for parasitological examination, DNA extraction, reaction preparation and DNA amplification. We have strong evidence that no contamination occurred, and the nucleotide sequences obtained were not identical.
Hookworm ITS-1-5.8S-ITS-2 locus was recovered in 75% (3/4) of the hookworm-positive samples analysed (fig 1b). The 719 bp sequences were identical and presented 100% similarity with two distinct Necator americanus: isolates G5-H4 and I-1 (GenBank accession numbers LC088287 and AB793527, respectively).
In the present study, Kato–Katz thick smears were stored for 7 years before processing and DNA extraction. The results indicate that faeces preserved under the cellophane cover in the Kato–Katz slides are a potential DNA source for studies in molecular parasitology. This can significantly reduce logistical difficulties in the field in terms of obtaining, preserving, transporting and initial processing of samples. Obtaining parasitic DNA, not only from soil-transmitted helminths but also from Schistosoma mansoni, which is still endemic in some Brazilian regions, is hampered by the need for cryopreservation of faecal samples obtained in remote regions, as the preservatives traditionally used to fix faecal specimens, such as formalin, greatly hamper procedures. The technique applied here may allow for an increase in the number of samples analysed and large-scale analysis in genetic, epidemiological and drug-resistance studies.
Author ORCIDs
L.H. Jaeger https://orcid.org/0000-0001-8031-7742.
Acknowledgements
The authors thank the Program for Technological Development in Tools for Health-PDTIS/FIOCRUZ for use of its facilities.
Financial support
This work was supported by the Oswaldo Cruz Foundation (Fiocruz), Brazilian National Council for Scientific and Technological Development (CNPq), and the Federal Agency for Support and Evaluation of Graduate Education (CAPES).
Conflict of interest
None.
Ethical standards
The study was approved by the Evandro Chagas Clinical Research Institute Committee for Ethics on Research (license no. 0011.0.009.000-3).




