Introduction
Accurate identification of genetic diversity within germplasm collections is essential for establishing and managing appropriate breeding programmes. The characterization of germplasm and analyses of population and conservation genetics based on molecular markers have gained importance due to the speed and quality of the generated data (de Vicente et al., Reference de Vicente, Guzmán, Engels and Ramanatha Rao2005; Govindaraj et al., Reference Govindaraj, Vetriventhan and Srinivasan2015). Microsatellites (SSRs) are highly informative molecular markers (Oliveira et al., Reference Oliveira, Pádua, Zucchi, Vencovsky and Vieira2006; Vieira et al., Reference Vieira, Santini, Diniz and Munhoz2016) that have been developed for many plant species and are currently used in molecular breeding, germplasm evaluation, genetic diversity, genome mapping, hybridization and evolutionary studies (Gupta and Varshney, Reference Gupta and Varshney2000; Govindaraj et al., Reference Govindaraj, Vetriventhan and Srinivasan2015; Tambarussi et al., Reference Tambarussi, Veasey, Menezes, Ibañes, Lombardi and Vencovsky2017).
In general, only a few sets of microsatellite markers are described in the literature for many species, despite the advantages they offer. Thus, there is a clear need to expand the available sets of these highly informative genetic markers, which can inform the effective management of species, improve germplasm resources, accelerate breeding programmes (Bajay et al., Reference Bajay, Zucchi, Kiihl, Batista, Monteiro and Pinheiro2011; Amorim et al., Reference Amorim, Silva, Ferreira, Amorim, Santos, Vilarinhos, Santos, Souza Júnior and Miller2012), and produce more accurate results in population genetics studies. As such, Tambarussi et al. (Reference Tambarussi, Menezes, Ibañes, Antiqueira, Dequigiovanni, Moreno, Ferraz, Zucchi, Veasey and Vencovsky2016) developed eight microsatellite markers for Cattleya walkeriana for both commercial and conservation interests and new studies have been developed using these markers. However, to improve accuracy and increase the pool of available microsatellites, the present study provides 22 new polymorphic loci for future genetic studies on these endemic and endangered Brazilian orchids.
Experimental
Total genomic DNA was extracted from fresh leaves collected from a single plant of C. walkeriana (voucher number: 23052) from the Orchid Germplasm Collection of the Genetics Department (ESALQ/USP), University of São Paulo, Piracicaba, São Paulo, Brazil, using the protocol described by Doyle and Doyle (Reference Doyle and Doyle1990). A microsatellite-enriched genomic library was constructed following the protocol of Billotte et al. (Reference Billotte, Lagoda, Risterucci and Baurens1999), with the same criteria as that described in Tambarussi et al. (Reference Tambarussi, Menezes, Ibañes, Antiqueira, Dequigiovanni, Moreno, Ferraz, Zucchi, Veasey and Vencovsky2016). Thirty-two specimens of C. walkeriana were analysed (voucher numbers: 23052a, 23052b, 23055, 2663, 85, 129RP, 100, 214, 907d, 905, 906, 882, 879, 887, 136RP, 1628, 12260, 132RP, 89, 128RP, 23189, 2409, 135RP, 1627, 907, 130RP, 88, 23207, 23193, 23068, 125RP and 2709). We also genotyped 12 accessions of Cattleya loddigesii (voucher numbers: 31077, 31073, 33626, 34026, 34029, 34030, 34033, 34034, 34044, 34046, 34050 and 34060) and six accessions of Cattleya nobilior (voucher numbers: 01, 02, 2354d, 30982, 30999 and 5652) to test the transferability of the microsatellite primers. All specimens of the three-species come from different populations.
Polymerase chain reactions (PCR) were performed in a final volume of 10 µl, using 5 µl of HotStarTaq Master Mix 2× (QIAGEN, Hilden, Germany), with 8 µM of M13 tail and reverse primers, 2 µM of forward primer according to Schuelke (Reference Schuelke2000) and 50 ng/μl of DNA. PCR cycling conditions were: initial denaturation at 95°C for 5 min, followed by 30 amplification cycles (95°C for 1 min, 1 min at the specific annealing temperature of each primer pair (online Table S1), 72°C for 1 min) and a final elongation step at 72°C for 7 min. Subsequently, 1 µl of the PCR product of each set of primers was added to 10 µl solution of formamide and GeneScan LIZ 500 dye Size Standard following the manufacturer's instructions. After solution denaturation at 95°C for 5 min, the PCR products were placed on an ABI 3130xl DNA analyser (Applied Biosystems) for automated capillary electrophoresis. For data analysis, we used the software GeneMapper v.5 (Applied Biosystems).
Genetic diversity was determined based on the number of alleles per locus (k) and observed (H o) and expected (H e) heterozygosity for each locus, and as an average across all loci. To test for linkage disequilibrium between pairwise loci, we used 1000 Monte Carlo permutations (alleles among individuals) and a Bonferroni correction (95%, α = 0.05). These estimates were calculated using the FSTAT version 2.9.3.2 program (Goudet, Reference Goudet1995). Null allele frequencies for each locus was estimated using the program Micro-Checker 2.2.3 (van Oosterhout et al., Reference van Oosterhout, Weetman and Hutchinson2006), and verified using a Maximum-Likelihood approach implemented in the INEST version 2.1 program (Chybicki and Burczyk, Reference Chybicki and Burczyk2009). The power to exclude the first parent (when none of the relatives are known) (![]() $$P_{1 ^{\circ} {\rm parent}}$$) and allelic combined non-exclusion probability of identity (I) were estimated using Cervus 3.0.3 (Marshall et al., Reference Marshall, Slate, Kruuk and Pemberton1998).
$$P_{1 ^{\circ} {\rm parent}}$$) and allelic combined non-exclusion probability of identity (I) were estimated using Cervus 3.0.3 (Marshall et al., Reference Marshall, Slate, Kruuk and Pemberton1998).
Discussion
All primer pairs amplified for C. walkeriana, but four and five primers showed no amplification for C. loddigesii and C. nobilior, respectively. We found 118 alleles (average of 5.1) for C. walkeriana, 73 alleles (average of 4.2) for C. loddigesii, and we identified 45 alleles (average of 2.6) for C. nobilior (Table 1; online Table S2). For C. walkeriana, H o and H e ranged from 0.100 to 0.966 and from 0.095 to 0.900, respectively. For C. loddigesii and C. nobilior, the average H o was 0.479 and 0.531, and H e was 0.562 and 0.478, respectively (Table 1; online Table S2).
Table 1. Results of initial primer screening in specimens of C attleya walkeriana
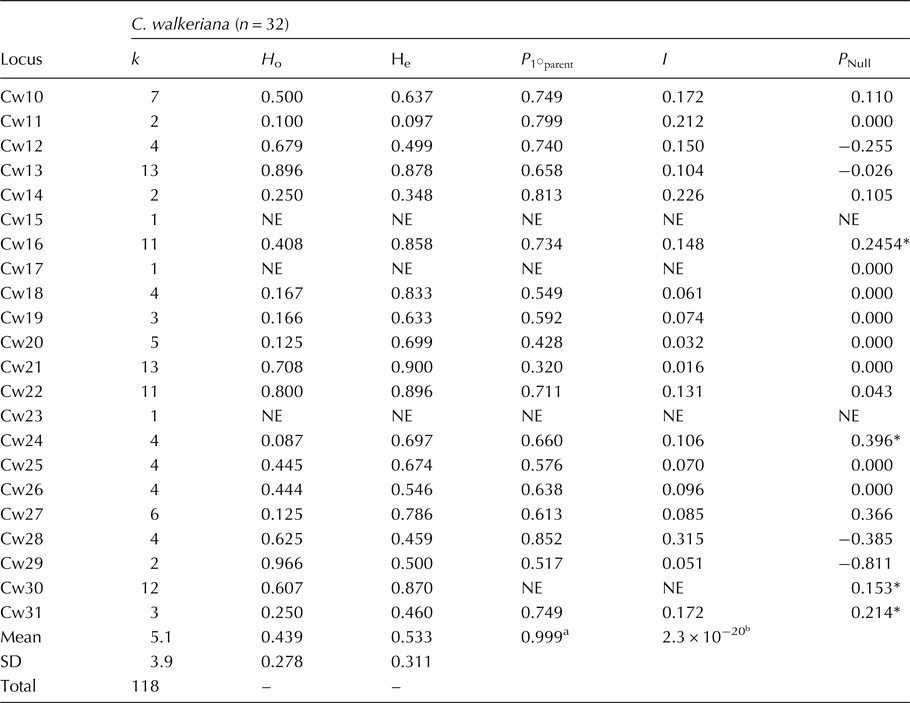
H e, expected heterozygosity; H o, observed heterozygosity; n, sample size for each population; k, the number of alleles per locus. *P Null, van Oosterhout estimate for the frequency of null alleles at each locus. NE, not estimated. ![]() $$P_{1 ^{\circ} {\rm parent}}$$ is the power to exclude the first parent (when no parent is known). I, combined non-exclusion probability of identity.
$$P_{1 ^{\circ} {\rm parent}}$$ is the power to exclude the first parent (when no parent is known). I, combined non-exclusion probability of identity.
a Combined exclusion probability.
b Combined estimation.
Analysis using the Micro-Checker software revealed null alleles at four loci (Cw16, Cw24, Cw30 and Cw31) for C. walkeriana. However, due to the small sample size for C. lodigesii and C. nobilior, Micro-Checker could not perform the analysis. However, null alleles were not observed at any locus for the three-studied species using a Maximum-Likelihood approach. No linkage disequilibrium was detected in any pair of loci. Only one locus was deemed unsuitable for C. walkeriana due to low polymorphism or lack of amplification, while for C. lodigesii and C. nobilior, eight and 10 loci, respectively, are unsuitable (Table 1; online Table S2). All other loci are appropriate for genetics studies on these orchid species. The ![]() $$P_{1 ^{\circ} {\rm parent}}$$ (0.999) and I (2.3 × 10−20) values indicate that the use of these loci are suitable for future parentage studies. The combined non-exclusion probability of identity (I) supports the use of these loci for identity analyses of these Cattleya species (Table 1; online Table S2).
$$P_{1 ^{\circ} {\rm parent}}$$ (0.999) and I (2.3 × 10−20) values indicate that the use of these loci are suitable for future parentage studies. The combined non-exclusion probability of identity (I) supports the use of these loci for identity analyses of these Cattleya species (Table 1; online Table S2).
These SSRs loci are suitable for genetic studies and can assist the investigation of possible interspecific crosses between species. They can also be used to support in situ and ex situ conservations, breeding programmes, genetic diversity and structure, mating system and gene flow of C. walkeriana and their related species.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262117000193.
Acknowledgements
We thank the Laboratório de Reprodução e Genética de Espécies Arbóreas (LARGEA, ESALQ/USP) for providing the physical support necessary to complete this work and to ‘Professor Paulo Sodero Martins’ Orchid Germplasm Collection of the Genetics Department (ESALQ/USP), University of São Paulo, Piracicaba, São Paulo, Brazil that provided the samples of species. RV, APS and MIZ were supported by a National Counsel of Technological and Scientific Development (CNPq) research fellowship.



