Introduction
Reproductive interference is one of the major factors mediating species exclusion among insects (Reitz & Trumble, Reference Reitz and Trumble2002). Negative sexual interactions between species caused by reproductive interference, such as misdirected courtship and heterospecific mating, occur when species with incomplete mate recognition systems overlap in the same habitat, and often results in the reduction of fitness for at least one of the interacting species (Gröning & Hochkirch, Reference Gröning and Hochkirch2008). Effects of reproductive interference can take various forms, such as reduced opportunities to mate with conspecifics, sexual harassment and damage to the female genitalia (Hochkirch et al., Reference Hochkirch, Gröning and Bücker2007; Gröning & Hochkirch, Reference Gröning and Hochkirch2008). Reproductive interference is often asymmetric, where the fitness of one species is affected more than another and continued sexual interactions between them can lead to exclusion of the inferior species, a process known as sexual exclusion (Gröning & Hochkirch, Reference Gröning and Hochkirch2008; Kishi et al., Reference Kishi, Nishida and Tsubaki2009). Theoretical and experimental studies suggest that because reproductive interference is characterized by positive frequency dependence, it is far more likely to cause species exclusion than the density dependence of resource competition (Kuno, Reference Kuno1992; Hochkirch et al., Reference Hochkirch, Gröning and Bücker2007; Kishi et al., Reference Kishi, Nishida and Tsubaki2009). Because biological invasions often involve species exclusion, the role of reproductive interference in biological invasions has received increasing attention (Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007; Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ; Wang et al., Reference Wang, Crowder and Liu2012; Luan et al., Reference Luan, De Barro, Ruan and Liu2013).
The whitefly Bemisia tabaci (Gennadius) has a global distribution and enormous genetic diversity (De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011; Boykin et al., Reference Boykin, Armstrong, Kubatko and De Barro2012). Recent molecular phylogenetic analyses and crossing experiments between genetic groups show that B. tabaci is a species complex containing at least 35 morphologically indistinguishable species (De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011; Hu et al., Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011a ; Boykin et al., Reference Boykin, Armstrong, Kubatko and De Barro2012; Liu et al., Reference Liu, Colvin and De Barro2012). In this species complex, two cryptic species, Middle East-Asia Minor 1 (MEAM1) and Mediterranean (MED), formerly referred to as the ‘B biotype’ and ‘Q biotype’ respectively, have risen to global prominence in the past 20 years (De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011; Sun et al., Reference Sun, Xu, Luan and Liu2011). Since the late 1980s, MEAM1 has invaded at least 54 countries from its origin in the MEAM region, and since early 2000s, MED has invaded at least ten countries from its origin in the MED region (De Barro & Ahmed, Reference De Barro and Ahmed2011; De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011; McKenzie et al., Reference McKenzie, Bethke, Byrne, Chamberlin, Dennehy, Dickey, Gilrein, Hall, Ludwig, Oetting, Osborne, Schmale and Shatters2012). In both the regions of their origin and those they have invaded, MEAM1 and MED have caused considerable losses to a range of major crops through direct feeding and virus transmission (Crowder et al., Reference Crowder, Horowitz, Breslauer, Rippa, Kontsedalov, Ghanim and Carrière2011; De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011).
Although molecular phylogenetic analyses indicate that MEAM1 and MED went through allopatric evolution (De Barro & Ahmed, Reference De Barro and Ahmed2011; Boykin et al, Reference Boykin, Armstrong, Kubatko and De Barro2012), field surveys in the past 20 years in various parts of the world indicate that both species often occur in sympatry (Crowder et al., Reference Crowder, Horowitz, Breslauer, Rippa, Kontsedalov, Ghanim and Carrière2011; Hu et al., Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011a ; Pan et al., Reference Pan, Chu, Ge, Wang, Wu, Xie, Jiao, Liu, Yang, Yang, Su, Xu and Zhang2011; Tsueda & Tsuchida, Reference Tsueda and Tsuchida2011; Saleh et al., Reference Saleh, Laarif, Clouet and Gauthier2012). In many cases, MEAM1 and MED have been found to co-occur on the same plots of crops or weeds (Crowder et al., Reference Crowder, Horowitz, Breslauer, Rippa, Kontsedalov, Ghanim and Carrière2011; Hu et al., Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011a ; Pan et al., Reference Pan, Chu, Ge, Wang, Wu, Xie, Jiao, Liu, Yang, Yang, Su, Xu and Zhang2011). Laboratory observations also show that the two species occur on similar parts of the plants (Muniz et al., Reference Muniz, Nombela and Barrios2002). Thus behavioural interactions between MEAM1 and MED in the field are likely to be widespread, and may contribute substantially to their interactions.
Studies with an integrated approach using behavioural observations, population cage experiments, field sampling and heuristic modelling have shown that reproductive interference plays an important role in the exclusion of indigenous whitefly species by MEAM1 (Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007; Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a , Reference Crowder, Sitvarin and Carrière b ; Luan & Liu, Reference Luan and Liu2012; Luan et al., Reference Luan, De Barro, Ruan and Liu2013) or MED (Wang et al., Reference Wang, Crowder and Liu2012). Laboratory observations on populations of MEAM1 and MED from Spain (Pascual, Reference Pascual2006), Israel (Elbaz et al., Reference Elbaz, Lahav and Morin2010) and Japan (Tsueda & Tsuchida, Reference Tsueda and Tsuchida2011) indicated that when MEAM1 and MED feed in mixed populations on the same plants, reductions in fecundity and/or proportions of females in progeny usually occur in MED, but not in MEAM1, suggesting asymmetric reproductive interference in favour of MAEM1. However, these studies did not examine the behavioural mechanisms underlying the reproductive interference and the population consequences. A more detailed study on the reproductive interference between MEAM1 and MED has been conducted by Crowder et al. (Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a , Reference Crowder, Sitvarin and Carrière b ) with populations of the two species from Arizona, USA, and suggests a possible link between asymmetric reproductive interference and the sexual exclusion of MED by MEAM1.
Although a few studies have been conducted on the reproductive interference between MEAM1 and MED, more detailed research on this aspect with the populations of the two invasive species in China is critically needed. This is because: (1) rapid and widespread exclusion of MEAM1 by MED has been occurring in many localities in China since the arrival of MED around 2003, and this exclusion seems to be contradictory to the laboratory observation of stronger reproductive interference with MED by MEAM1, as outlined above; (2) the genetic structure of either MEAM1 or MED shows considerable diversity (e.g., De Barro & Ahmed, Reference De Barro and Ahmed2011; Wang et al., Reference Wang, Luan, Li, Su, Xia and Liu2011; Chu et al., Reference Chu, Hu, Gao, Zhao, Nichols and Li2012), and different populations of the same species may behave differently; and (3) our studies on reproductive interference between MEAM1 (or MED) and indigenous whiteflies (Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007; Luan & Liu, Reference Luan and Liu2012; Luan et al., Reference Luan, Xu, Lin, Zalucki and Liu2012; Wang et al., Reference Wang, Crowder and Liu2012) suggest that detailed observations of mating behaviour help to discern the mechanisms underlying reproductive interference and exclusion. Therefore, we conducted species exclusion experiments, behavioural observations and population modelling to investigate the species exclusion between MEAM1 and MED in China, as well as the behavioural mechanisms underlying species exclusion. We show that for the populations of the two invasive whiteflies in China, MEAM1 has an intrinsic, stronger capacity for excluding MED than vice versa, and this capacity is associated with asymmetric behavioural interactions favouring MEAM1.
Materials and methods
Whiteflies and plants
The population of MEAM1 (mtCO1 GenBank accession no. AJ332557) used in this study was collected from cucumber, Cucumis sativus L. in September 2009, in Rui'an, Zhejiang; the population of MED (mtCO1 GenBank accession no. GQ371165) was collected from pepper, Capsicum annuum L., in Ningbo, Zhejiang, China. The populations were maintained in separate climatic cubicles on cotton, Gossypium hirsutum (Malvaceae) cv. Zhe-Mian 1793, a host plant suitable to both MEAM1 and MED (Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013). The purity of each of the two populations was monitored every three generations using the random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) technique with the primer H16 (5′-TCTCAGCTGG-3′) (De Barro & Driver, Reference De Barro and Driver1997). Newly emerged whitefly adults from each population were used in the species exclusion experiments and behavioural observations (Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013).
Cotton plants (cv. Zhe-Mian 1793) used in the experiments were cultivated singly in potting mix (a mixture of peat moss, vermiculite, organic fertilizer and perlite in a 10:10:10:1 ratio by volume) in 1.5-L pots in whitefly-proof glasshouses where temperature and humidity were controlled at 24–30 °C and 50–70% RH, and natural lighting was supplemented with 14 h artificial lights during the daytime. All experiments used plants at the 5–7 fully expanded true leaf stage and were conducted at 27±1 °C, L14:D10 and 70±10% RH.
Species exclusion experiments
We conducted population cage experiments to observe changes in relative abundance as well as sex ratios in mixed populations of MEAM1 and MED. Three experiments were conducted with different initial relative abundance of each of the two species. In the first experiment, we conducted three treatments (fig. 1): (i) MEAM1+MED in mixed population, ten replicates; (ii) MEAM1 alone, two replicates; and (iii) MED alone, two replicates. The experiment was conducted using steel-framed insect rearing cages (55 cm×55 cm×55 cm). The two treatments of MEAM1 and MED single populations served as controls. To initiate each experimental unit (cage), newly emerged adults were introduced to a cage containing two cotton plants. In treatment ‘MEAM1+MED’, the two plants in each cage were inoculated with ten females and ten males of MEAM1 and ten females and ten males of MED; in ‘MEAM1 alone’, the two plants in each cage were inoculated with 20 females and 20 males of MED; and, in ‘MED alone’, the two plants in each cage were inoculated with 20 females and 20 males of MED. The plants were watered as necessary.

Fig. 1. Changes of relative proportions and sex ratios of MEAM1 and MED in mixed cohorts of the two cryptic species on cotton in the laboratory which were initiated with 50% MEAM1 and 50% MED. (A) mean percentages of MEAM1 individuals in cohorts of mixed population of ‘MEAM1+MED’, cohorts of MEAM1 alone, and cohorts of MED alone, respectively; (B) mean percentages of females of MEAM1 in the cohorts of mixed population of ‘MEAM1+MED’ and in the cohorts of MEAM1 alone, respectively; (C) mean percentages of females of MED in the cohorts of mixed population of ‘MEAM1+MED’ and in the cohorts of MED alone, respectively. Error bars indicate standard errors. In (B) and (C), different letters to the right of the four mean values on the same day indicate significant differences (P<0.05). Note that raw percentage data are shown but that statistical analyses were performed on arcsine-square root transformed data.
For both MEAM1 and MED, the development time from egg to adult emergence takes 23–25 days on average under the tested host plant and temperature conditions (Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013). Thus, every 25 days over a 125-day period, in the MEAM1+MED treatment, 100 whitefly adults were sampled from each experimental unit (cage) where individual whiteflies were drawn from each of all plant leaves and identified to gender and species; in each cage of the two control treatments, 30 adults were examined by RAPD-PCR for their species identity and 100 individuals were sexed. To avoid overcrowding and maintain the population in each cage, after each sampling of the adults, the older plant of the two in each cage was cut and taken out with all the eggs and nymphs on it, and a new clean plant was added (note that in the first sampling, the two plants in each cage were of the same age, we removed the plant that appeared to have less leaf area). Sampling ended when no individuals of one of the species were detected in the samples.
The protocols for initiating the second and third experiments were the same as those of the first experiment, except that relatively lower proportion of MEAM1 was used in the mixed cohorts of the two species. In the second experiment, the two plants in each experimental unit (cage) of the mixed population were inoculated with five females and five males of MEAM1 and 15 females and 15 males of MED, with six replicates (fig. 2). In the third experiment, the two plants in each experimental unit (cage) of the mixed population were inoculated with two females and two males of MEAM1 and 18 females and 18 males of MED, with six replicates (fig. 3). Then, for both experiments, every 75 days (about three generations) 100 whitefly adults were sampled from each cage where individual whiteflies were drawn from each of all plant leaves and identified to gender and species.

Fig. 2. Changes of relative proportions of MEAM1 and MED in mixed cohorts of the two species on cotton in the laboratory which were initiated with 25% MEAM1 and 75% MED.

Fig. 3. Changes of relative proportion of MEAM1 and MED in each of six mixed cohorts of the two species on cotton in the laboratory which were initiated with 10% MEAM1 and 90% MED.
Behavioural observations
We used the video recording system of Ruan et al. (Reference Ruan, Luan, Zang and Liu2007) to observe the mating behaviour and copulation events of adults caged on plant leaves (Sun et al., Reference Sun, Xu, Luan and Liu2011). One female and one male MEAM1 or MED adult was supplemented with one male of the same or the other species. Three treatments were conducted for each of the two species (tables 1 and 2). Newly emerged adults of various intra- and inter-species treatments were caged on the lower surface of plant leaves, and their movement and behaviour were observed and recorded continuously for 72 h. The events of courtship and copulation, as well as behavioural interactions between individuals of the same or different species, were determined by viewing the tapes on a television set or a computer screen.
Table 1. Courtship events and interactions: behavioural elements that caused changes in events of copulation in MEAM1 when a pair of MEAM1 ♂+♀ was supplemented with one ♂ of MEAM1 or MED during the first 3 days after emergence.

The data in the table are mean±SEM, and means on the same row followed by different letters indicate significant differences (P<0.05).
Table 2. Courtship events and interactions: behavioural elements that caused changes in events of copulation in MED when a pair of MED ♂+♀ was supplemented with one ♂ of MED or MEAM1 during the first 3 days after emergence.

The data in the table are mean±SEM, and means on the same row followed by different letters indicate significant differences (P<0.05).
The courtship and mating behaviour of B. tabaci has been described in detail (Perring & Symmes, Reference Perring and Symmes2006; Zang & Liu, Reference Zang and Liu2007). In replay of the tapes, we determined the following behavioural events: (i) copulation: a successful copulation event between a male and female; (ii) courtship: a male and a female positioned parallel to each other with their bodies in contact; (iii) interference: an intruding male interfered with the courtship or copulation of a male and a female; (iv) successful interference without displacement: an event of interference that resulted in immediate, early ending of courtship or copulation, but the intruding male did not replace the earlier male; and (v) successful interference leading to displacement: an event of interference that resulted in replacement of the first male in courting by the intruding male. With the recording of these behavioural elements, we were able to calculate the number of uninterrupted events of courtship, i.e. events of courtship that ended naturally without experiencing any interference. Uninterrupted events of courtship could lead to copulation or could end without copulation.
For treatments where each replicate had only female and males of the same species, we did not need to distinguish individual males, and thus we viewed tapes on a television set. For treatments where each replicate had one female with males from both MEAM1 and MED, we need to identify each male to species at each behavioural event, and thus we viewed the tapes on a computer installed with the Motic Images Advanced 3.2 system (Motic China Group Co. Ltd, Xiamen, China). The techniques for distinguishing individual males with the aid of the Motic Images Advanced 3.2 system on a computer screen are reported in detail in Luan & Liu (Reference Luan and Liu2012). Briefly, the actual lengths of the two males in each experimental unit were measured and recorded before they were released for the observation, and they were then identified by the difference in their relative body length, i.e. one was longer than the other.
Effect of mating interactions on fecundity and progeny sex ratio
In parallel with behavioural observations, we also examined the progeny production by MEAM1 or MED using ten intra- and inter-species treatments (fig. 4c–f). Newly emerged adults of the ten treatments were caged on the lower surface of plant leaves, and left to mate and oviposit for 5 days before being discarded. All eggs on the plants were reared for 30 days for them to develop to adults, and all progeny adults were then collected and sexed.
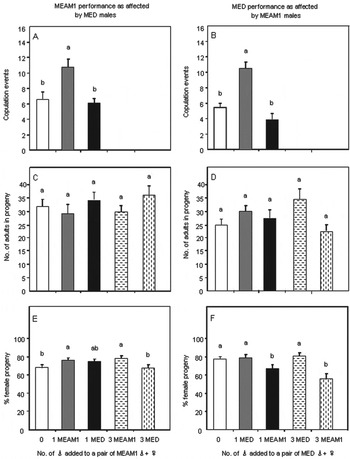
Fig. 4. Changes in the mean number of copulation events during the first 72 h after emergence and production of progeny for the first 5 days after emergence when a pair of MEAM1 ♂+♀ was supplemented with one or three ♂ of the MEAM1 and MED (A, C and E), or when a pair of MED ♂+♀ was supplemented with one or three ♂ of the MED or MEAM1 (B, D and F). Ten to 27 replicates were conducted for each of the ten treatments, and error bars indicate standard errors. In each of the six diagrams different letters above bars indicate significant differences (P<0.05). Note that in Panels E and F raw percentage data are shown but that statistical analyses were performed on arcsine-square root transformed data.
Modelling species exclusion
To determine if the observed mating behaviours could predict patterns of species exclusion observed in the population cage experiments, we used the stochastic simulation model of Crowder et al. (Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ); see also Wang et al. (Reference Wang, Crowder and Liu2012). The full model has been described (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ), and we only summarize it here. In the model, female behaviour was simulated on a per-individual basis, with each female was courted once per time step (1 h) until she was mated. The probability of a courtship ending in copulation P success =P intra ×P cop , where P intra is the probability of an intra-species courtship, and P cop is the probability of copulation in intra-species courtships. Values for these parameters were based on the behavioural experiments. Each courtship was simulated by drawing a random number from a uniform distribution between 0 and 1, which was compared to the observed probability values for P intra and P cop . If either random number was greater than the observed probability, the courtship ended before mating; otherwise, the courtship ended in copulation. Both mated and unmated females laid eggs, with female fecundity peaking at age 2 and 3 days and declining thereafter (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ). Unmated females laid only male progeny, whereas the progeny sex ratio of mated females depended on whether a competing species was present. The model had functions for adult and immature development on a daily basis, and was written in Visual Basic (Microsoft, 2002).
We conducted simulation with two models to evaluate whether variation in mating interactions between MEAM1 and MED could predict the patterns of species exclusion observed in the experiments. Respectively, the two models were (i) behaviour model – simulations with variation in mating behaviour only, and (ii) control model – mating behaviour was the same for both species. The control model simulated a scenario where no behavioural interactions occurred, whereas the behaviour model simulated variation between MEAM1 and MED in mating behaviour due to reproductive interference. In both models, we assumed that parameter values for life history were the same for MEAM1 and MED. We ran simulations for the same time frame as the experiments, with each of the three starting values for the proportion of MEAM1 used to initiate the experiments. The number of stochastic simulations used in data analysis matched the number of experimental replicates in each case (ten replicates with 50% MEAM1 initially, six replicates with 25% or 10% MEAM1 initially). We also conducted a second set of simulations, with 100 stochastic model runs per set of conditions, to explore the likelihood of species exclusion based on the initial proportion of MEAM1. For these simulations, models were run until one species excluded the other.
Data analysis
For behavioural observations, the data of different treatments was analysed using one-way analysis of variance (ANOVA); and, when a significant effect was detected at the P<0.05 level, the means were compared using a least significant differences (LSD) test. For species exclusion experiments, the percentage of females in the same generation with different treatments was analysed using one-way ANOVA; and, when a significant effect was detected at the P<0.05 level, the means were compared using a LSD test. For modelling species exclusion experiments between MEAM1 and MED, we used repeated measures ANOVA to determine if the proportion of MEAM1 over time was affected by model (control or behaviour), time and the interaction between model and time. A separate modelling was conducted for each set of initial conditions (i.e., the proportion of MEAM1). All proportion data were transformed by arcsine square root before the analysis. All statistical analyses were done using the statistical software, STATISTICA (version 6.1) (StatSoft Inc., 2003).
Results
Species exclusion experiments
In the first experiment, the relative abundance of MEAM1 increased steadily in the first two generations, from 50 to 60%, and then increased rapidly and reached 100% by the 5th generation, i.e. MED was completely excluded by MEAM1 (fig. 1a). No inter-species contamination occurred in the two control treatments of MEAM1 and MED (fig. 1a).
In the meantime, the population sex ratio of the two species in the three treatments also experienced some changes. All cohorts of each of the two species used to initiate experiments had 50% females. Percentages of females of both MEAM1 and MED increased to 60–70% in the first generation. Thereafter, in MEAM1 the percentage of females remained at that level in both the MEAM1 alone and MEAM1+MED treatments; whereas in MED the percentages of females remained at that level in the MED alone treatment but decreased significantly on the third and fourth generations in the MEAM1+MED treatment (fig. 1b and c).
In the second experiment, the relative abundance of MEAM1 increased steadily from 25% at the start to 35.6% in the third generation, and then to 54.8% in the sixth generation (fig. 2). In the third experiment, two contrasting outcomes occurred: in four of the six replicates, the relative abundance of MEAM1 increased steadily from 10% at the start to 25–42% in the sixth generation, whereas in two of the replicates, the relative abundance of MEAM1 decreased steadily from 10% at the start to 0% in the sixth generation, i.e. MEAM1 was completely displaced by MED (fig. 3).
Behavioural interactions
The mean numbers of copulation events between MEAM1♂ and MEAM1♀ increased when a MEAM1 male was added but did not change when a MED male was added (table 1; F 2, 37=7.1, P<0.01). The mean number of courtships between MEAM1♂ and MEAM1♀ increased significantly when a MEAM1 male was added (F 2, 37=34.4, P<0.01); the mean numbers of uninterrupted courtship events between MEAM1♂ and MEAM1♀ also increased when a MEAM1 male was added (F 2, 37=21.7, P<0.01). The mean numbers of uninterrupted courtship between MEAM1♂ and MEAM1♀ per MEAM1♂ did not differ significantly among the three treatments (F 2, 37=0.52, P=0.60); however, the mean percentages of uninterrupted courtship events leading to copulation between MEAM1♂ and MEAM1♀ were significantly reduced when either a MEAM1 male or a MED male was added (F 2, 37=20.2, P<0.01).
The mean number of copulation events between MED♂ and MED♀ increased significantly when a pair of MED adults was supplemented with a MED male; whereas the mean number of copulation events between MED♂ and MED♀ appeared to be reduced when one MEAM1 male was added although the difference was not significant (table 2; F 2, 37=17.1, P<0.01). The mean numbers of courtship events between MED♂ and MED♀ increased significantly when a MED male was added (F 2, 37=32.8, P<0.01); the mean numbers of uninterrupted courtship events between MED♂ and MED♀ also increased significantly when a MED male was added (F 2, 37=20.9, P<0.01). The mean numbers of uninterrupted courtship between MED♂ and MED♀ per MED♂ did not differ significantly between the three treatments (F 2, 37=2.55, P=0.09). However, the mean percentages of events of uninterrupted courtship leading to copulation between MED♂ and MED♀ did not change significantly when a MED male was added but decreased significantly when a MEAM1 male was added (F 2, 37=32.2, P≤0.01).
Comparison of other events in mating interactions between the two species indicates some apparent differences (tables 1 and 2). Compared to the situation of one species alone, in the presence of males of the other species, females in both species reduced acceptance of uninterrupted courtships with males of their own species for copulation. However, the reduction in MED was greater (from 79 to 24%, table 2) than that in MEAM1 (from 81 to 45%; table 1). In addition, while the frequencies of interference with courtships of the other species were similar in the two species, MEAM1 males achieved success of interference in 21% (0.6/2.9) of the events but MED males never achieved success (0/2.7). MEAM1 males also made more frequent attempts in courting females of the other species than MED males (12.4 versus 6.1). These results indicate that when the two species co-occur, compared with MED, MEAM1 females have higher acceptance of courtships leading to copulation and MEAM1 males have stronger capacity to interfere with courtships of the other species.
We also observed the durations of pre-copulatory courtships and copulations. Both MEAM1 and MED significantly increased duration of copulation in the presence of males of the other species but did not increase duration of copulation in the presence of males of its own species (table 3).
Table 3. Durations of pre-copulatory courtship and copulation between females and males in MEAM1 and MED in response to an additional male of the same or the other species.

The data in the table are mean±SEM, and means of the three treatments of A or B on the same column followed by different letters indicate significant differences (P<0.05).
Effect of mating interactions on fecundity and progeny sex ratio
When a pair of MEAM1 ♀+♂ were supplemented with males of either MEAM1 or MED, the mean numbers of adults in the progeny did not change significantly (fig. 4c; F 4, 95=0.67, P=0.61); percentages of females in the progeny either increased when MEAM1 males were added or did not change significantly when MED males were added (fig. 4e; F 4, 95=3.91, P<0.05). When a pair of MED ♀+♂ were supplemented with males of MED or MEAM1, the mean numbers of adults in the progeny did not change significantly (fig. 4d; F 4, 66=1.33, P=0.27); percentages of females in the progeny either did not change significantly when MED males were added or decreased significantly when MEAM1 males were added (fig. 4f; F 4, 66=4.29, P<0.05).
Modelling species exclusion
We derived values of various parameters for the models based on the behavioural experiments as well as the experiments examining the effect of mating interactions on fecundity and progeny sex ratio (table 4). For ‘Progeny sex ratio of mated females’, the alternative parameter values 0.69 and 0.51 for MEAM1 and MED were their respective mean proportions of female progeny of the two species observed in the experiments examining the effect of mating interactions on fecundity and progeny sex ratio. In the two treatments of ‘1MEAM1♂+1MEAM1♀+1MED1♂’ and ‘1MEAM1♂+1MEAM1♀+3MED1♂’, each with ten replicates, the overall mean proportion of female progeny of MEAM1 was 0.69, whereas in the two treatments of ‘1MED♂+1MED♀+1MEAM1♂’ and ‘1MED♂+1MED♀+3MEAM1♂’, each with ten replicates, the overall mean proportion of female progeny of MED was 0.51 (fig. 4e and F). For ‘female behaviour (copulation with male of intra-species per hour)’, the alternative parameter values 0.085 and 0.054 for MEAM1 and MED were calculated from the data recorded in the behavioural observation. In the treatment ‘1MEAM1♂+1MEAM1♀+1MED1♂’, each MEAM1♀ copulated on average 6.1 times in 72 h and thus 0.085 times h−1 (table 1), whereas in the treatment ‘1MED♂+1MED♀+1MEAM1♂’, each MED♀ copulated on average 3.9 times in 72 h and thus 0.054 times h−1 (table 2). For ‘Male behaviour (% courtships initiated with MEAM1 female)’, each MEAM1 female on average did courtships for 11.7 times with MEAM1 male and 6.1 times with MED male in 72 h (66% versus 34%; table 1). For ‘male behaviour (% courtships initiated with MED female’, each MED female on average did courtships for 12.4 times with MEAM1 male and 7.2 times with MED male (63% versus 37%; table 2).
Table 4. Parameter values for modelling species exclusion between MEAM1 and MED: behavioural traits were assumed to be the same for the two species in the control simulations, whereas alternative parameter values were assumed based on experimental observations on the performance of the two species.

For each set of initial conditions, the control model failed to predict the observed changes in the relative abundance of MEAM1 over time (data type effect: P≤0.012 in all cases, table 5, fig. 5). Furthermore, over time these models diverged from the observed data (data type×time effects: P<0.05 in all cases, table 5). In contrast, the models that incorporated mating behaviour predicted changes in MEAM1 frequency over time that did not differ significantly from the observed patterns (data type effect: P≥0.10 in all cases, table 5, fig. 5). In one case, the data type×time interaction was significant (MEAM1 initially=50%), as the observed proportion of MEAM1 increased slightly faster than the model predicted (P=0.0002, table 5, fig. 5). However, when the initial % of MEAM1 was 25% or 10%, the models predicted the observed changes precisely over time (P≥0.17, table 5, fig. 5). As observed in the experiments, models initiated with 50% or 25% MEAM1 always led to exclusion of MED (fig. 6). Also similar to the experiments, when models were initiated with 10% MEAM1, MED was excluded in 62% of simulations (fig. 6). Thus, behavioural traits appeared to significantly drive exclusion of MED by MEAM1, and predicted the rate at which exclusion occurred across a variety of initial conditions.

Fig. 5. The proportion of MEAM1 over time that was observed in experiments or predicted with two models (control and behaviour).

Fig. 6. The proportion of simulations incorporating variation in mating behaviour where MED was excluded by MEAM1, with varying initial proportion of MEAM1. Also shown is the proportion of population cages where MED was excluded (or appeared to be in the process of being excluded) by MEAM1.
Table 5. Results of repeated measures ANOVA comparing observed with simulated data from two models on the proportion of MED in the mixed population of MEAM1 and MED during the cage experiments.

Discussion
Although studies on mating behaviour and behavioural interactions have been reported for populations of MEAM1 and MED in USA and Israel (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a , Reference Crowder, Sitvarin and Carrière b , Reference Crowder, Horowitz, Breslauer, Rippa, Kontsedalov, Ghanim and Carrière2011), our report represents the first investigation of this aspect on populations of the two invasive species in China, a region where the two species have shown a different pattern of competitive displacement in the field from that recorded in USA and Israel. The combined data of the three experiments on species exclusion (figs 1–3) show that in this enclosed, homogenous condition, MEAM1 is able to exclude MED in <20 generations as far as the initial relative abundance of MEAM1 is above 10%, demonstrating a stronger intrinsic capacity of MEAM1 for competition when interacting with MED. The reduction in proportion of females in MED indicates that the MEAM1's ability to exclude MED is associated with asymmetric reproductive interference favouring MEAM1 (fig. 4). The role of this behavioural mechanism in mediating the species exclusion was further shown by the modelling on species exclusion (fig. 5). Interestingly, in two of the six replicates where the initial proportion of MEAM1 was 10%, MED displaced MEAM1 in six generations. This latter result indicates that when the mixed population of the two species reaches this relative proportion between them, the trajectory of their interactions may be affected by priority effects, allowing MED to exclude MEAM1 in some cases when MED is initially very common. The validity of this observation was also shown by the modelling on species exclusion (fig. 6).
Our behavioural observations showed intensive mating interactions between MEAM1 and MED (tables 1–3). As has been observed in previous studies on MEAM1 and MED (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ; Elbaz et al., Reference Elbaz, Lahav and Morin2010; Sun et al., Reference Sun, Xu, Luan and Liu2011), females and males of the two species exhibit frequent courtships, although they do not, or very rarely, copulate. Our data revealed two major differences in mating interactions between the two species: (1) compared with MED, females of MEAM1 exhibit higher acceptance to courtships of its own males leading to copulation when males of the other species are present, and (2) MEAM1 males have stronger capacity for interference with the courtships of the other species than MED males (table 6). Higher acceptance of courtships by females leading to copulation in MEAM1 than in MED was also reported for the populations of the two species from USA (Crowder et al., Reference Crowder, Sitvarin and Carrière2010b ), and a stronger capacity of MEAM1 males for behavioural interference than MED males was also reported for the populations of the two species from Spain (Pascual, Reference Pascual2006), Israel (Elbaz et al., Reference Elbaz, Lahav and Morin2010), USA (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ) and Japan (Tsueda & Tsuchida, Reference Tsueda and Tsuchida2011). The agreements among these studies indicate that the differences in behavioural traits between MEAM1 and MED revealed in this study are present in many populations of the two species around the world.
Table 6. Summary of the differences in mating behavioural interactions between MEAM1 and MED as well as those between MEAM1 and indigenous (Asia II 3 and Australia; for species designation see De Barro et al., Reference De Barro, Liu, Boykin and Dinsdale2011) whitefly species.

Asymmetric mating interactions have been reported between MEAM1 and a number of indigenous whitefly species (Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007; Luan & Liu, Reference Luan and Liu2012; Luan et al., Reference Luan, Xu, Lin, Zalucki and Liu2012, Reference Luan, De Barro, Ruan and Liu2013; Wang et al., Reference Wang, Crowder and Liu2012). Comparison of the interactions between the two invasive species MEAM1 and MED, as reported here as well as in some earlier studies (e.g., Crowder et al., Reference Crowder, Sitvarin and Carrière2010b ) and those between MEAM1 and indigenous species show differences in many aspects (table 6). Note that the descriptions on the relative performance of MEAM1 versus Asia II 3/Australia were derived in a manner similar to that for MEAM1 versus MED in the present study. For example, behavioural observations showed that in response to the presence of additional males of the other species, MEAM1 males significantly increased their courtship events whereas the two indigenous species did not change their behaviour (Liu et al., Reference Liu, Colvin and De Barro2007; Luan et al., Reference Luan, De Barro, Ruan and Liu2013), and thus the description of ‘Yes versus No’ in the table. Remarkably, in the seven behavioural traits outlined in table 6, MEAM1 and MED show similarity in five when they interact, whereas MEAM1 and indigenous species show similarity in only one when they interact. This difference between the two categories of species interactions indicates that the degree of asymmetry in mating interactions is weaker between MEAM1 and MED than that between MEAM1 and indigenous species. The weaker asymmetry in mating interactions between MEAM1 and MED seems to agree with its milder adverse effects on the progeny sex ratio of MED compared with its effects on the progeny sex ratio of the indigenous species (e.g., compare fig. 4 here with fig. 3 of Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007). At the population level, under similar conditions, the exclusion of MED by MEAM1 is progressing considerably slower than that of indigenous species by MEAM1 (e.g., compare figs 1–3 here with Fig. S3 of Liu et al., Reference Liu, De Barro, Xu, Luan, Zang, Ruan and Wan2007).
Although the asymmetry in mating interactions between MEAM1 and MED is relatively weaker than that between MEAM1 and indigenous species, its role in mediating species exclusion between the two species is still dramatic (figs 1–3, 5 and 6). This asymmetry will certainly offer MEAM1 advantages in its competition with MED in the field, as has been shown for the two species in Israel (Crowder et al., Reference Crowder, Horowitz, Breslauer, Rippa, Kontsedalov, Ghanim and Carrière2011). The question then arises: why has MED been excluding MEAM1 in many localities in China (Chu et al., Reference Chu, Wan, Zhang and Brown2010; Hu et al., Reference Hu, De Barro, Zhao, Wang, Nardi and Liu2011a ; Pan et al., Reference Pan, Chu, Ge, Wang, Wu, Xie, Jiao, Liu, Yang, Yang, Su, Xu and Zhang2011; Rao et al., Reference Rao, Luo, Zhang, Guo and Devine2011; Shen et al., Reference Shen, Du, Ren and Qiu2011; Guo et al., Reference Guo, Rao, Luo, Zhang and Gao2012), a scenario that also appears to be occurring in Korea (Park et al., Reference Park, Jahan, Song, Lee, Lee, Choi, Lee, Kim, Lee and Lee2012) and Japan (Iida et al., Reference Iida, Kitamura and Honda2009; Tsueda & Tsuchida, Reference Tsueda and Tsuchida2011)?
To address this question, we must consider that interspecific interactions are affected by a range of intrinsic and environmental factors, and the roles of competition and reproductive interference must be evaluated in the context of the environments where the species interact (Reitz & Trumble, Reference Reitz and Trumble2002). In the last 10 years in China, widespread and very frequent application of a range of insecticides such as neonicotinoids has been used against whiteflies. The MED populations in China have significantly higher levels of resistance than those of MEAM1 to nearly all commonly used insecticides (Rao et al., Reference Rao, Xu, Luo, Zhang, Jones, Devine, Gorman and Denholm2012; Yuan et al., Reference Yuan, Wang, Zhou, Du, Zhang and Wang2012; Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013). The substantially higher levels of resistance to insecticides in MED, coupled with the widespread and heavy input of insecticides has been shown to play a major role in assisting MED in its exclusion of MEAM1 in China (Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013). A similar result has been shown in population cage experiments involving these two species in Israel, where MEAM1 excluded MED when insecticides were not applied, but the opposite result (MED excluded MEAM1) occurred in the presence of insecticide use (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ). The results of this study also show that the degree of asymmetry in mating interactions between MEAM1 and MED in favour of MEAM1 is weaker than that between MEAM1 and indigenous species (table 6), and once the relative proportion of MEAM1 drops to 0.1 or lower MED may win the competition and displace MEAM1 (figs 3, 5 and 6).
In contrast to the situations recorded in China's mainland, Japan and Korea, MED has not been detected in outdoor agriculture in the USA (McKenzie et al., Reference McKenzie, Bethke, Byrne, Chamberlin, Dennehy, Dickey, Gilrein, Hall, Ludwig, Oetting, Osborne, Schmale and Shatters2012) and Taiwan (Hsieh et al., Reference Hsieh, Chiang and Ko2011) until now, although the whitefly arrived in the USA and Taiwan at about the same time as that for China's mainland, Japan and Korea. The reasons for this disparity are unclear. In the USA, one study on host plant use of MED indicated that the MED population there has an apparently poorer capacity to adapt to field crops than the populations of MED in Japan or China (Iida et al., Reference Iida, Kitamura and Honda2009; Hu et al., Reference Hu, Dennehy, Ni, Zhao, Nichols and Li2011b ; Tsueda & Tsuchida, Reference Tsueda and Tsuchida2011). The other possible reason could be the difference in insecticide input between China's mainland and USA. In some states of the USA such as Arizona, successful development and implementation of whitefly-IPM programmes in the past 15 years have reduced the use of insecticides against whitefly to very low levels, and the reduction in insecticide application in turn has helped the rich fauna of whitefly natural enemies to recover and exert substantial control on the whitefly populations (Naranjo & Ellsworth, Reference Naranjo and Ellsworth2009). As the data of this study, Sun et al. (Reference Sun, Liu, Qin, Xu, Li and Liu2013) and Crowder et al. (Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a ) have shown, under scenarios of no insecticide application, MED is unable to co-exist with MEAM1, primarily due to reproductive interference by the latter. However, when insecticides are used, MED can exclude or coexist with MEAM1 (Crowder et al., Reference Crowder, Horowitz, De Barro, Liu, Showalter, Kontsedalov, Khasdan, Shargal, Liu and Carrière2010a , Reference Crowder, Horowitz, Breslauer, Rippa, Kontsedalov, Ghanim and Carrière2011; Sun et al., Reference Sun, Liu, Qin, Xu, Li and Liu2013).
In summary, our study demonstrates that for the populations of the two invasive whiteflies in China, MEAM1 has a stronger capacity for excluding MED than vice versa, and this capacity is associated with asymmetric reproductive interference favouring MEAM1. Although the asymmetry in reproductive interference is likely to offer MEAM1 advantages over MED in the field, the multiple interspecific differences between the two species and the environmental heterogeneity, part of which is related to human manipulation, may promote niche partitioning and co-existence of the two species across China, or even favour MED in some seasons/regions. Ultimately, linking detailed laboratory and population cage studies with long-term field datasets may provide the key to predicting patterns of species exclusion or coexistence across landscapes and design better management strategies.
Acknowledgements
Financial support for this study was provided by the China Agriculture Research System (CARS-25-B-08), the National Natural Science Foundation of China (31272104) and the USDA-AFRI NIFA Fellowship Program (2011-67012-30718).














